INTRODUCTION
Cervical cancer is a type of cancer that occurs in the uterine cervix, and it is the second most common cancer and fourth leading cause of death among women worldwide (Singh et al., 2023) and high-risk human papillomavirus (HR-HPV) is the most important pathogenic factor. Persistent HR-HPV infection causes abnormal proliferation of cervical squamous epithelial cells and can lead to cervical intraepithelial neoplasia.
DNA methylation refers to the binding of a methyl group to cytosine, occurring predominantly at CpG sites where cytosine and guanine are adjacent. Since CpG sites are abundant in promoter sequences of genes, methylation at CpG sites is one of the crucial factors influencing gene transcription (Costello & Plass, 2001).
In unicellular organisms, DNA methylation serves as a defense mechanism against the intrusion of exogenous genetic material. Conversely, in multicellular organisms, particularly during ontogenesis, DNA methylation assumes a pivotal role in modulating gene expression. Recent research findings have elucidated that the methylation patterns of genes associated with pathological conditions can exert dual effects on gene expression, either promoting or inhibiting it (Phillips, 2008; Moore et al., 2013; Kaplun et al., 2022). Genes subject to DNA methylation influence diverse biological pathways including the cell cycle, cellular proliferation, apoptosis, and metastasis. Notably in oncogenesis, DNA methylation is implicated in tumorigenesis by epigenetically silencing tumor suppressor genes through hypermethylation and activating oncogenes via hypomethylation (Baylin et al, 1998; Ehrlich, 2002).
Homeobox A11 (HOXA11), a member of the homeobox (HOX) gene family, encodes a transcription factor that plays a pivotal role in embryonic development and cellular differentiation (Li et al., 2019). The HOX genes are highly conserved across species and are critical regulators of morphogenesis, controlling the spatial and temporal expression of genes involved in developmental processes (Bhatlekar et al., 2018). HOXA11, in particular, is essential for the proper development of the reproductive tract, as evidenced by its involvement in the differentiation and function of the uterine stroma and myometrium.
Recent studies have suggested that aberrant expression and epigenetic modifications of HOXA11 are implicated in various cancers (Cui et al., 2015; He et al., 2024). Methylation-induced silencing of HOXA11 has been observed in cancerous tissues, leading to the hypothesis that HOXA11 functions as a tumor suppressor gene. This hypothesis is supported by findings demonstrating that the restoration of HOXA11 expression inhibits cell proliferation and induces apoptosis in cancer cell lines. The mechanistic pathways through which HOXA11 exerts its tumor-suppressive effects remain to be fully elucidated. In this study, we aim to investigate the role of HOXA11 in cervical cancer in depth, examining its methylation status and the consequent effects on cellular proliferation and apoptosis. We seek to provide foundational data on the molecular mechanisms of HOXA11 function and thereby offer insights into its potential as a tumor marker and a therapeutic target for cancer treatment.
MATERIALS AND METHODS
Patients and normal cervical scrapes were obtained from the Chungnam National University Hospital. Genomic DNA was isolated from cervical scrapes using the QIAmp DNA Mini kit (cat. no. 51304; Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. Genomic DNA was chemically modified with sodium bisulfite using an EZ DNA Methylation Gold kit (cat. no. D5006; Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. Bisulfite-converted DNA was purified and eluted with an elution buffer using a Zymo-Spin IC column (Zymo Research).
To assess methylation status of HOXA11 gene, quantitative bisulfite-pyrosequencing (Dejeux, 2009) was performed. Specific bisulfite polymerase chain reaction (PCR) and pyrosequencing primers were designed to analyze 243 bp of CpG island including three CpG dinucleotides sites of HOXA11 gene using PyroMark Assay Design Software v. 2.0 (Qiagen). The following primers were used: forward, 5′-AGTAAGTTTATGG GAGGGGGATT-3’; reverse, 5’-Biotin-CCCCCATACAACATACTTATACTCA-3’; sequencing primer, 5’-TAGTTTA GGGTATTTTTTATTTAT-3’. Briefly, 20 ng of bisulfite-treated DNA was amplified in a 25 μL reaction with primer set and Taq polymerase (Enzynomics, Daejeon, Korea). PCR amplification was run for 40 cycles with an optimal annealing temperature.
Pyrosequencing was performed using a PyroGold kit and a PyroMarK ID Q96 instrument (Qiagen) following the manufacturer’s instructions. Each CpG site was assigned a percentage (%) of methylation by evaluating C/T ratio as methylation index. The average % of methylation across three CpG sites was obtained. Methylated non-CpG cytosines were used as internal controls to check the fidelity of bisulfite conversion. All pyrosequencing reactions included samples without any DNA template as negative controls.
HeLa and HT-3 cell lines were obtained from the American Type Culture Collection (ATCC). All cell lines were incubated at 37°C in a humidity 5% CO2 incubator, and cultured in each recommended medium (HeLa, Dulbecco’s Modified Eagle’s Medium, Sigma-Aldrich, St. Louis, MO, USA; HT-3, McCoy’s 5A Medium, Welgene, Gyeongsan, Korea) supplemented with 10% fetal bovine serum (FBS, Welgene) and 1% Amphotericin B/Streptomycin/Penicillin (Gibco, Grand Island, NY, USA). Cultures were replaced every 2–3 days, and all cells were cultured in 80%–90% confluence. HOXA11 expression vector was constructed by cloning full length HOXA11 into pcDNA3.1 vector using EcoR I and Xho I restriction enzymes. Restriction enzymes were purchased from Roche. Transfection was performed using the FUGENE HD (Roche, Mannheim, Germany) according to the manufacturer’s instructions.
Transiently transfected HeLa and HT3 cells grown in 24-well plates were trypsinized and harvested at 1 day intervals for 3 days. Cells were counted with a hemocytometer, and each experiment was repeated three times in triplicate wells.
HeLa and HT3 cell lines were seeded into 4 well chamber slide and transiently transfected with pcDNA3.1-HOXA11. After 48 hours of transient transfection, TUNEL assay was performed using In Situ Cell Death Detection Kit (Roche Applied Science, Penzberg, Germany) in accordance with the manufacturer’s instruction. Briefly, phosphate buffered saline (PBS) washed cells were fixed in 4% paraformaldehyde pH (7.4) for 1 hour at room temperature followed by incubation in permeabilization solution (0.1% Triton X-100, 0.1% sodium citrate) for 2 minutes on ice. Cells were washed twice with PBS and added 50 μL of TUNEL reaction mixture to each sample and incubated in a humidified dark chamber for 1 hour at 37°C. The cells were washed three times with PBS and analyzed with a fluorescent microscope.
HeLa cells were seeded into 100 mm culture plates and transiently transfected with pcDNA3.1-HOXA11. After 48 hours of transient transfection, mitochondrial and cytosol fractionation were isolated using Mitochondrial isolation kit for mammalian cells (Theromo Fisher, Waltham, MA, USA) in accordance with the manufacturer’s instruction.
Cells were lysed and equal amounts of cell extracts (20 μg) were electrophoresed on 8%–10% SDS polyacrylamide gel, electro-transferred onto a nitrocellulose membrane, and probed with antibodies. All proteins were quantitated by using the Bradford assay. After blocking for 1 hour at room temperature with 5% Skim milk (Biopure, Cambridge, MA, USA), the membrane was incubated overnight at 4°C with primary antibody. After that, the appropriate secondary antibody was treated at room temperature for 1 hour, and detection was performed using the enhanced chemiluminescence system (Amersham, Piscataway, NJ, USA). The antibodies used were as follows. HoxA11 (sc-81288), cMyc (sc-40), Bax (sc-493), VEGF (sc-152), cytochrome C (sc-13156), GAPDH (sc-47724) were purchased from Santa cruz. cleaved PARP (#9541) was purchased from Cell Signaling (Danvers, MA, USA).
RESULTS
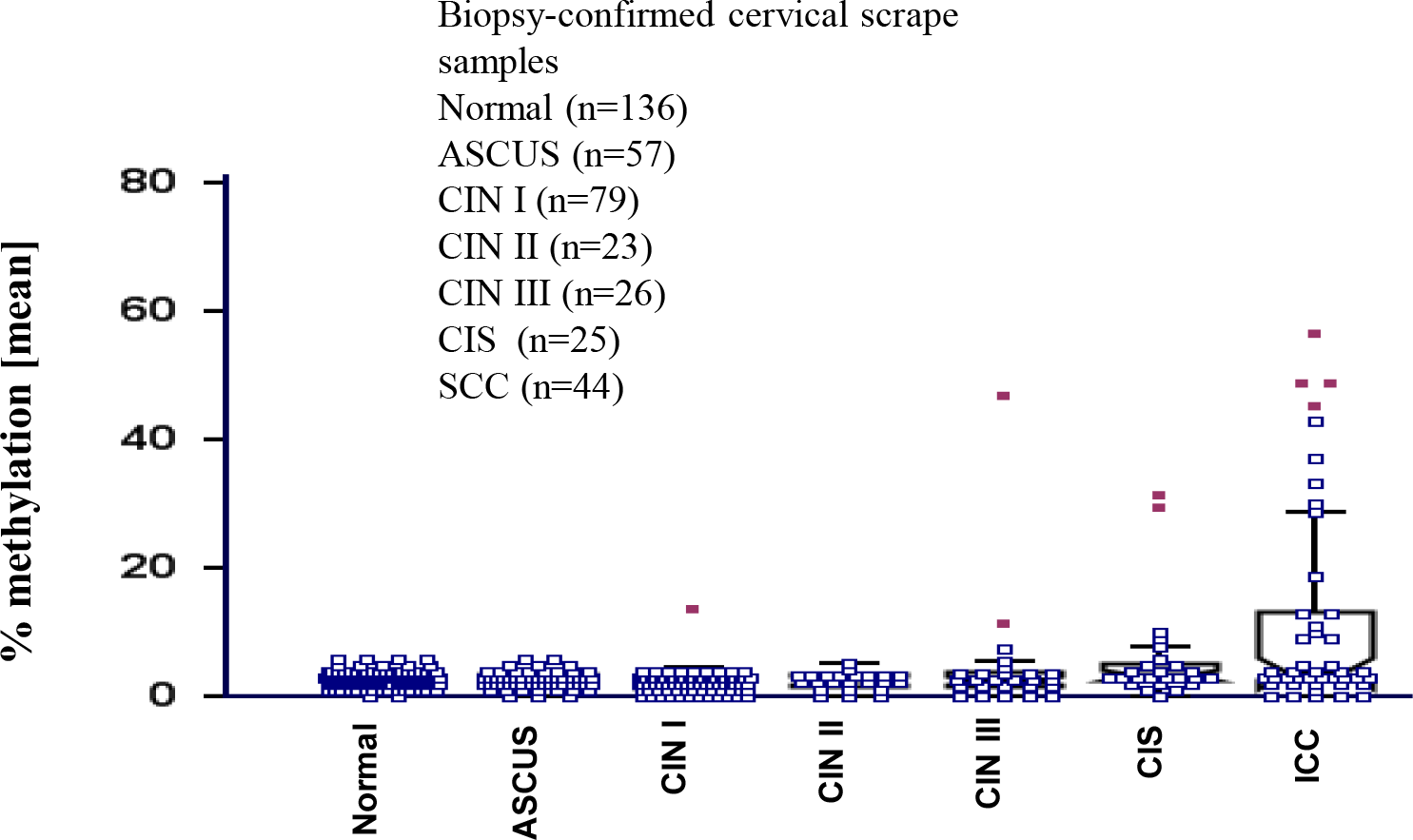
To identify epigenetically down-regulated genes in invasive cervical cancer (ICC), We first conducted a cDNA microarray to analyze the genes with altered expression in cervical cancer cell lines and HOXA11 gene was identified as a down regulated gene (data not shown). The methylation status of the HOXA11 promoter region was investigated using quantitative bisulfite-pyrosequencing methods across pathological states from normal to ICC scrapes (Fig. 1). The methylation levels of HOXA11 gene was significantly higher in ICC compared to those of healthy individuals (p<0.01). Aberrant hypermethylation of tested promoter region was more frequently observed in ICC (53.2% positive call, 23/44) compared to normal scrapes (8.1% positive call, 11/135). This indicates that aberrant hypermethylation of the promoter region is significantly more common in ICC scrapes compared to normal scrapes.
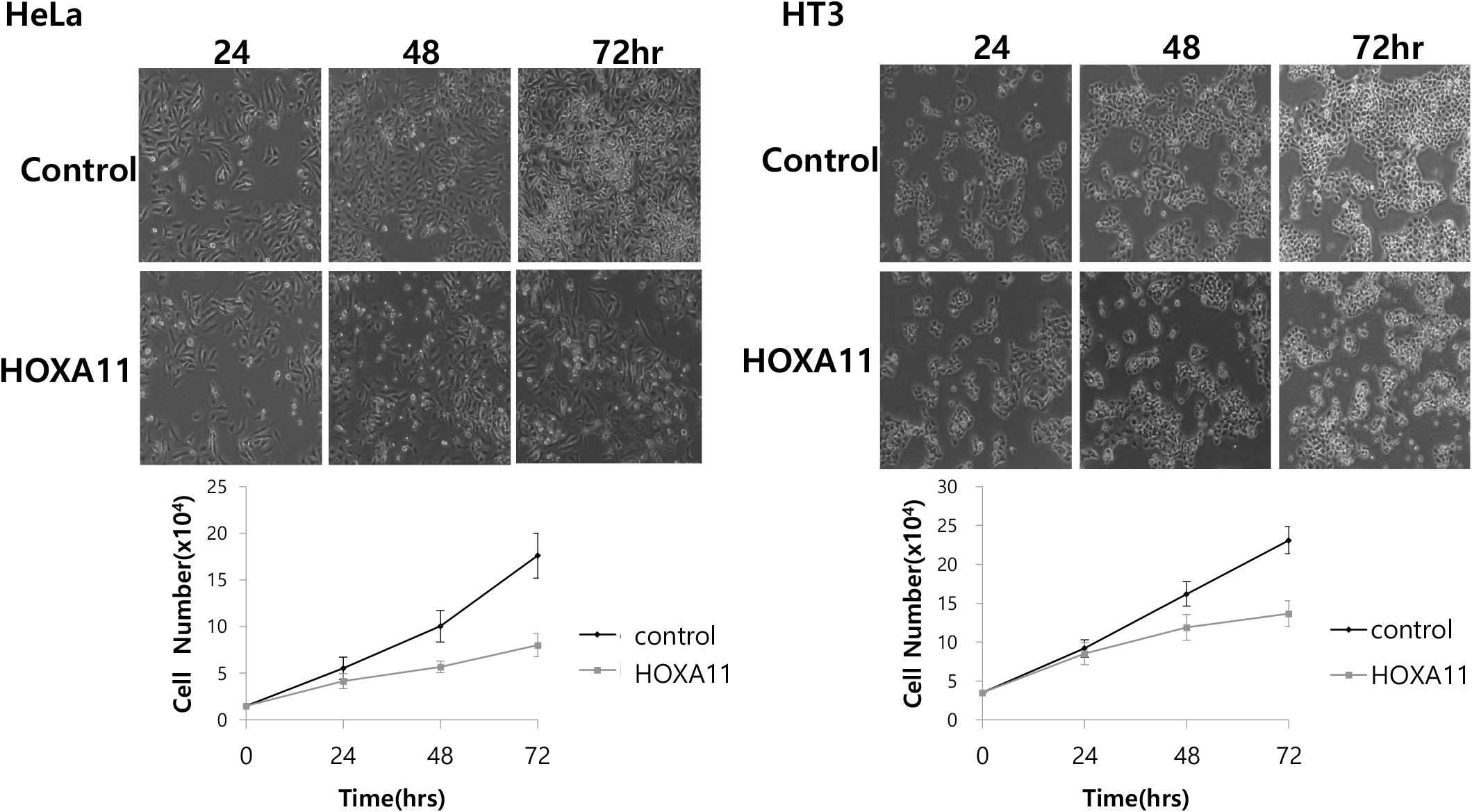
We next explored the effects of HOXA11 on cervical cancer cell growth by transfecting HeLa and HT3 cancer cells with HOXA11 expression vectors and monitored cell growth for 24–72 hrs. We found that the overexpression of HOXA11 inhibited cell growth by 50% in HeLa cells and by 30% in HT3 cells (Fig. 2). These results indicate that the overexpression of HOXA11, whose expression is inhibited by methylation, strongly suppresses the proliferation of cervical cancer cells.
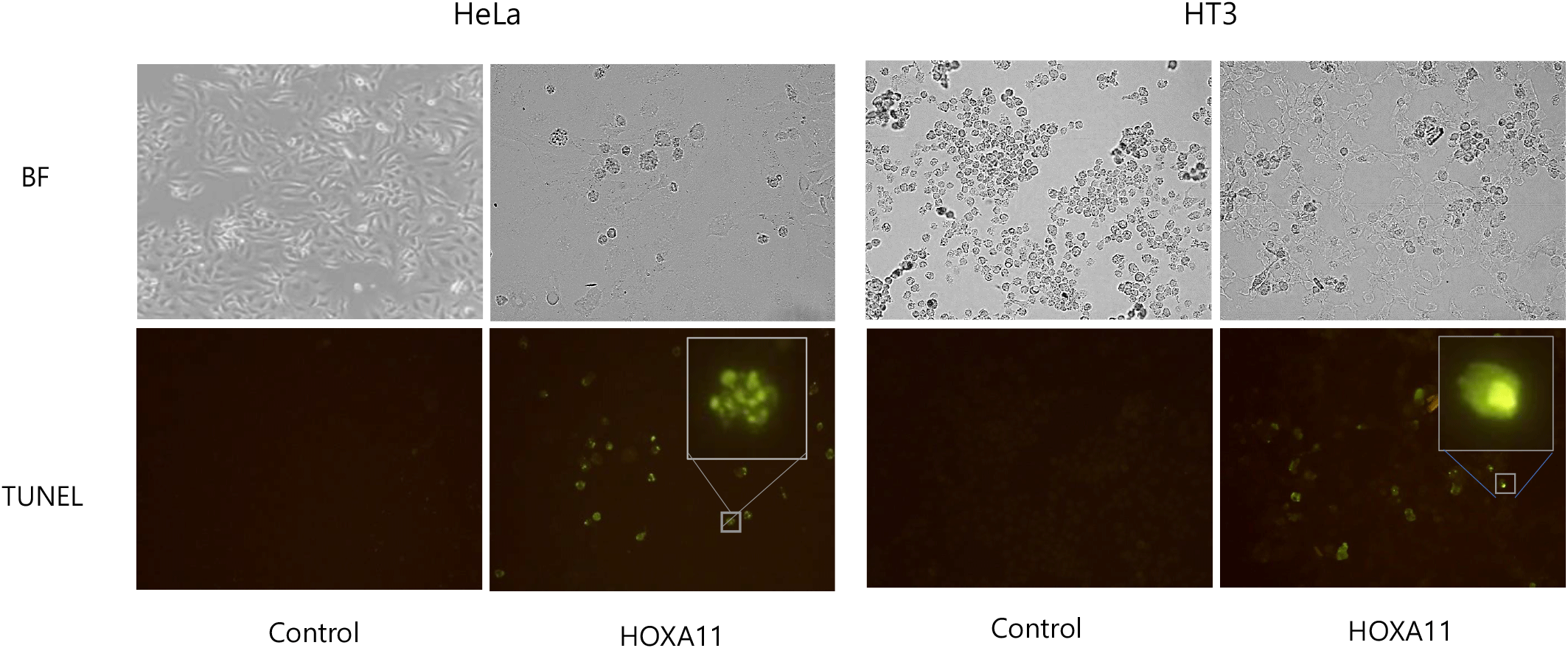
HOXA11 is down-regulated in cervical cancer cell lines and cancer scrapes. These results propose that down-regulation of HOXA11 might be cause of carcinogenesis in cervical cancer. Therefore, we examined whether over-expression of HOXA11 inhibits tumor cell growth by inducing apoptotic cancer cell death. We performed TUNEL assays and examined the morphology of nuclei in HeLa and HT-3 cells overexpressing control vector and HOXA11. HOXA11-overexpressing cells showed fragmented nuclei characteristic of apoptosis, whereas control cells contained normal nuclei (Fig. 3).
Cytochrome C is a key protein in the intrinsic pathway of apoptosis, which is the programmed cell death process. During apoptosis, cytochrome C is released from the mitochondria into the cytoplasm in response to pro-apoptotic signals. Based on this, the following experiment was conducted. After overexpressing HOXA11 in HeLa cell lines, the release of cytochrome C from the mitochondria to the cytoplasm was analyzed using Western blotting. The results showed that the concentration of cytochrome C in the cytoplasm increased significantly due to HOXA11 (Fig. 4A).
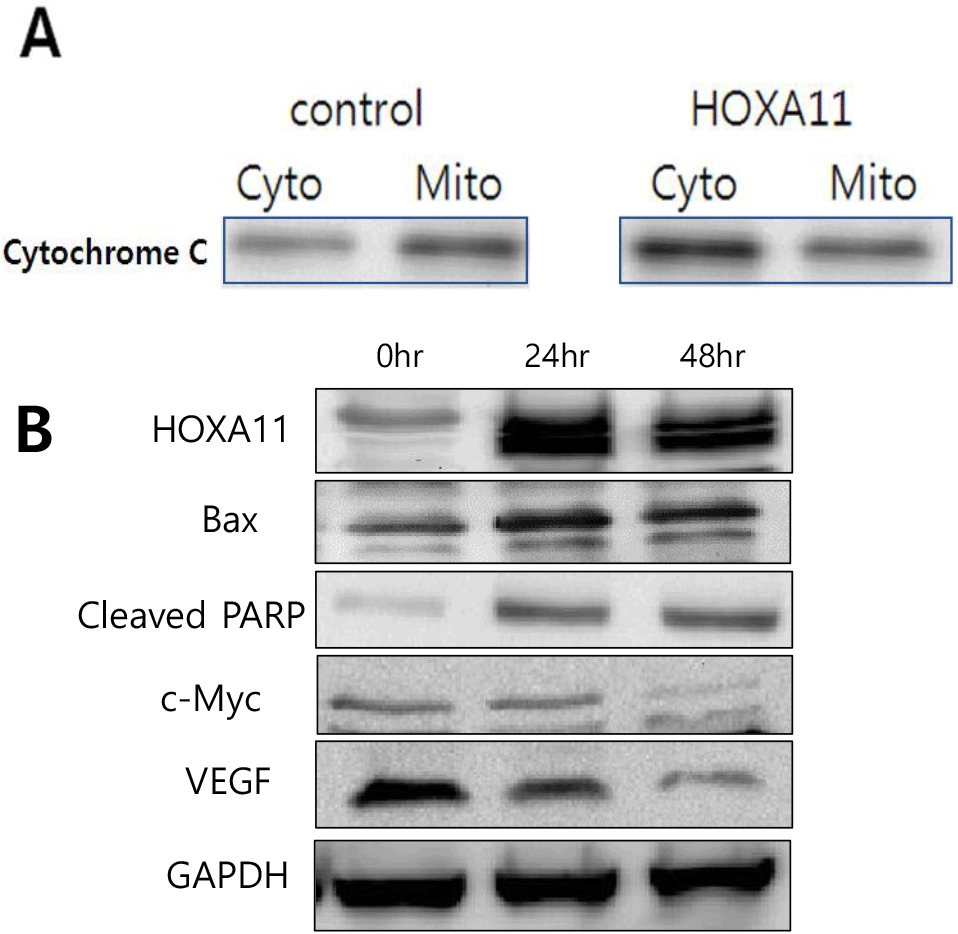
To further examine apoptosis, Bax (Bcl-2-associated X protein), a key proapoptotic protein that plays a crucial role in the intrinsic pathway of apoptosis and cleaved PARP as a marker of apoptosis were detected using Western blot analysis. HOXA11-overexpressing cells showed increased levels of Bax and cleaved PARP protein level in a time-dependent manner (Fig. 4B). Additionally we investigated the regulatory role of HOXA11 on the expression of c-Myc and VEGF, key regulators of cell proliferation and survival, respectively (Fig. 4B). Our findings revealed a notable inhibition in the expression levels of both c-Myc and VEGF when HOXA11 was overexpressed. These results mean that overexpression of HOXA11 induced high levels of apoptotic cell death in cervical cancer.
DISCUSSION
Epigenetic alterations play a major role in cancer development and DNA hypermethylation plays important roles in carcinogenesis by silencing tumor suppressor genes. Recent studies have reported that HOXA11 is hypermethylated in several cancers (Fiegl et al., 2008; Apostolidou et al., 2009; Li 2017).
The primary objective of this study was to elucidate the potential of HOXA11 as a diagnostic methylation marker and therapeutic target in uterine cancer. Our investigation commenced with a comprehensive analysis of HOXA11 gene expression using cDNA microarrays in cervical cancer cell lines, revealing a consistent and significant reduction in HOXA11 expression levels compared to normal controls (data not shown). This initial observation prompted further exploration into the epigenetic regulation of HOXA11 through promoter methylation analysis.
Subsequent investigation of HOXA11 promoter methylation status by quantitative bisulfite-pyrosequencing in samples obtained from both normal individuals and uterine cancer patients showed significant differences (Fig. 1). We found that HOXA11 promoter regions were markedly hypermethylated in cervical cancer scrapes, with methylation levels exceeding those observed in normal tissues by more than 53.2%. 53.2% difference may not sufficient to be used as a biomarker. But if we use cervical tissues the methylation percentage of HOXA11 should be elevated than scrape. Cervical tissues provide a more comprehensive representation of the molecular landscape within the uterine environment, encompassing both epithelial and stromal components. In contrast, scrapings typically yield cellular material predominantly from the epithelial surface, potentially limiting the depth of analysis and the ability to capture methylation changes occurring in deeper tissue layers or stromal cells. Therefore, while HOXA11 methylation levels showing a 53.2% increase in cancerous scrapings compared to normal scrapings provide initial insights, leveraging cervical tissues for methylation analysis could potentially enhance the discriminatory power of HOXA11 as a biomarker.
To assess the functional implications of HOXA11 dysregulation in cervical cancer, we conducted experiments involving the overexpression of HOXA11 in cervical cancer cell lines HeLa and HT3. Notably, heightened expression of HOXA11 resulted in a profound inhibition of cancer cell proliferation. This anti-proliferative effect was corroborated through TUNEL assay, which revealed morphological hallmarks of apoptosis in treated cells (Saraste & Pulkki, 2000), indicating that HOXA11-induced growth suppression occurs primarily through apoptotic cell death mechanisms. Mitochondrial apoptosis involves c-Myc activation triggering mitochondrial outer membrane permeabilization, releasing cytochrome c into the cytosol. Cytochrome c binds Apaf-1 to form the apoptosome, activating caspase-9. Caspase activation includes cleaving PARP, promoting DNA fragmentation and cell dismantling (Tait & Green, 2010; Glover et al., 2024). This process ensures controlled cell death essential for tissue health and development, with dysregulation implicated in diseases like cancer and neurodegeneration.
Further mechanistic insights into the apoptotic pathways modulated by HOXA11 overexpression highlighted its role in promoting mitochondrial apoptosis. Specifically, increased HOXA11 expression led to PARP cleavage, a hallmark of apoptosis, driven by elevated levels of c-Myc and augmented secretion of cytochrome C into the cytoplasm (Fig. 4A). These molecular changes suggest that HOXA11 exerts its anti-cancer effects by triggering intrinsic apoptotic pathways, thereby substantiating its potential as a therapeutic target in cervical cancer treatment strategies.
c-Myc and VEGF are important factors that respectively promote cell proliferation and survival, and they interact intricately with apoptosis (Kelly et al., 1983; Tai et al., 2012) c-Myc can induce apoptosis under specific conditions along with promoting cell growth, while VEGF provides survival signals that suppress apoptosis induced by c-Myc. Based on this, our study examined changes in the expression of c-Myc and VEGF regulated by HOXA11, revealing a significant inhibition in the expression of both c-Myc and VEGF. The downregulation of c-Myc is particularly significant in the context of cancer biology, as c-Myc is known to drive cell growth and proliferation. Its inhibition suggests a potential mechanism through which HOXA11 could exert tumor-suppressive effects by limiting excessive cell proliferation. Similarly, the suppression of VEGF expression has implications for angiogenesis and tumor vascularization. VEGF is crucial for promoting the formation of new blood vessels that supply nutrients and oxygen to tumors. Thus, the reduction in VEGF expression mediated by HOXA11 suggests a potential strategy to inhibit tumor growth and metastasis by limiting tumor angiogenesis. Our results underscore the intricate regulatory network involving HOXA11, c-Myc, and VEGF in cancer progression. The observed inhibition of c-Myc and VEGF expression supports the hypothesis that HOXA11 may act as a tumor suppressor by modulating key pathways involved in cell proliferation and angiogenesis.
In conclusion, the findings from this study provide compelling evidence supporting HOXA11 as both a diagnostic biomarker and a therapeutic candidate in cervical cancer. The distinct epigenetic signature of HOXA11 methylation in cancerous tissues underscores its potential clinical utility for early detection and prognosis. Moreover, the ability of HOXA11 to induce apoptotic cell death in uterine cancer cells highlights its therapeutic promise, suggesting avenues for the development of targeted therapies aimed at restoring HOXA11 function or enhancing its expression in cancerous tissues. Future directions should focus on validating these findings in clinical cohorts and exploring the translational potential of HOXA11-based interventions to improve outcomes for cervical cancer patients.







