INTRODUCTION
Lateral inhibition is known to be mediated by Delta-associated Notch signaling pathway (Kunisch et al., 1994). Within equivalent cells in proneural domain, signal-sending cells expressed Delta induce Notch receptor in neighboring cells, which become neuroblasts or sensory organ precursors (Bray, 1998). It is well known that Hairy Enhancer-of-split, a transcriptional repressor, is expressed by the transcriptional complex consist of notch intracellular domain (NICD), CBF1/Su(H)/Lag1 (CSL) and Mastermind (Mam) in neighboring cells (Bray, 1998). In neighboring cells of neural ectoderm, Ngn1 as a proneural gene is repressed by a Hairy Enhancer-of-split-related protein (Kim et al., 1997; Takke et al., 1999). In contrast to neighboring cells, Mind bomb (Mib) promotes ubiquitination and endocytosis of Delta with extracellular domain of Notch (NECD) in signal-sending cells (Itoh et al., 2003). Jagged have been identified as a ligand of Notch receptors (Lindsell et al., 1995). Zebrafish Jagged2 expressed in the domain located progenitor cells of motor neuron (pMN domain) activate Notch and maintain a population of proliferating progenitors in the p3 domain (Yeo & Chitnis, 2007). Jagged-mediated Notch signaling regulate the temporal generation of late-born neurons, such as secondary motor neurons (sMN) and GABAergic KA” neurons in the ventral spinal cord (Yeo & Chitnis, 2007). The gain-of-function study using Tg[hsp70-Mib:EGFP] showed that the majority of cells in the p3 domain differentiated into KA” neurons after heat-shock during late neurogenesis (Ryu et al., 2015). These observations imply that Jagged may also be a substrate for Mib. Our preliminary data showed that Jagged2 was ubiquitinated by Mib in P19 cells, but not cytosolic form of intracellular domain of zebrafish Jagged2 (J2ICD) (unpublished data). This observation implicate that an adaptor might be required for ubiquitinating membrane-bounded Jagged ICD by cytosolic Mib. By performing yeast-two hybrid screen using zebrafish Jagged2 ICD as a bait, zebrafish PEX1 was isolated and identified a potential adaptor between Mib and Jagged2 ICD.
Mutations of human PEX1 gene was initially identified in first complementation group (CG1) of the peroxisome biogenesis disorders (PBDs), a group of lethal autosomal-recessive diseases caused by defects in peroxisomal matrix protein import, with the simultaneous loss of multiple peroxisomal enzyme activities (Portsteffen et al., 1997; Reuber et al., 1997). Peroxisomes are metabolic organelles involved in β-oxidation of fatty acids, hydrogen peroxide-based respiration, and defense against oxidative stress (Brown & Baker, 2003). PEX1 have been studied as a member of AAA-ATPase family required for the formation and maintenance of peroxisomes, membrane-bound organelles with PEX6 (Geisbrecht et al., 1998; Judy et al., 2022). We have investigated the physiological meaning of interaction between Jagged2 and PEX1 in regulating the generation of late-born neurons in the p3 domain. Our study shows that zebrafish PEX1 deletion model phenocopies Jagged2 morphant, suggesting that ubiquitous expression of PEX1 may help to activate Jagged2-mediated Notch signaling pathway which maintain a population of proliferating progenitors in the ventral spinal cord.
MATERIAL AND METHODS
Zebrafish were maintained as described by Yeo & Chitnis (2007). The embryos were staged in accordance with the method of Kimmel et al. (1995). AB* wild-type and PEX1−/− mutant were used.
The sequences of zebrafish PEX1 (NM_001170835) was identified in GenBank. PCR was performed as described by Yeo & Chitnis (2007). PCR was performed using the primers as below; Pex1yt3FlagEco_FP01; 5’-t GAA TTC ATT TTG GAG CTG TCA TGG-3’, Pex1yt3Xho_RP01; 5-C CTC GAG TGC TAG CAG AAC ACA AAC-3’, Pex1ID_FP02; 5-GCC ATC CTG AAA GAC TGT ATC C-3’, Pex1ID_RP01; 5-TCC TGA CCA CAG CAG AAT TC-3’, J2SP_FP01; 5’-ATG TGG AAT TGT ATC AGG-3’, J2SP_RP02; 5’-TAA CAG CAG GCA CGC GAT-3’, J2TM_FP03; 5’-GTG GAT TAC CTG GTA CCT-3’, J2TM_RP04; 5’-CAC ACA GAC GAT GAC GCA-3’, J2ICD_FP05; 5’-TGG TGG ACC CGC AAG CGG-3’, J2ICD_RP06; 5’-TAC GCA GTG TTC TTT GTG-3’. PCR products were subcloned into the pGEM-T easy vector, pCS-MT or pCS-Flag vector and then verified by sequencing.
The Y2H screening was performed as described manufacturer (Clonetech, Terra Bella, CA, USA). The yeast strain was transformed with pDBT-J2ICD as bait. Positive clones were validated in the Y2H assay by checking for activation LacZ reporter genes. Isolated clones were determined by PCR and DNA sequencing.
The P19 cell line was grown in medium as described by Kong et al. (2018). COS cell line was grown in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with antibodies and 10% FBS. Cell lines were cultured at 37°C in a 5% CO2 incubator. Transient transfections were performed using FuGENE6 transfection reagent (Roche, Basel, Switzerland). Proteins samples for the Co-IP assay were prepared as follows. Transfected COS cells (10 cm dishes, 2×106 cells/dish, 1 dish per IP) were harvested 24-hour after transfection in 400 μL lysis buffer (20 mM HEPES [pH 7.5], 150 mM KCl, 0.5% NP-40 and 10% glycerol, and protease inhibitor), debris was removed by centrifugation. Then, 500 μL of the lysate was immunoprecipitated with the indicated antibodies and proteinA-coated beads. Immunoprecipitations were performed overnight at 4°C on a roller. The beads were washed three times with 500 μL wash buffer (150 mM NaCl, 1% Triton X-100, 10% glycerol, 50 mM Tris pH 8.0, 1 mM EDTA, 1 mM MgCl2) and proteins were eluted with 50 μL SDS loading buffer at 95°C for 4 min. Proteins were separated by SDS-PAGE and detected by immunoblotting.
Whole-mount in situ hybridization was performed as described by Yeo & Chitnis (2007). Antisense riboprobes were transcribed from cDNAs for zebrafish GAD1 and islet1 (Yeo & Chitnis, 2007). Photos were taken with a DIC microscope (Axioplan2; Carl Zeiss, Oberkochen, Germany).
Immuohistochemistry was performed as described by Yeo & Chitnis (2007). Primary antibodies used were as follows: anti-Myc antibody (Sigma-Aldrich, St. Louis, MO, USA), anti-Flag M2 antibody (Sigma-Aldrich) and zn5/DM-GRASP antibody (Developmental Studies Hybridoma Bank, Iowa City, IA, USA). Secondary antibodies used were as describe by Yeo & Chitnis (2007). Photo were taken with a confocal laser scanning microscope (LSM 510; Carl Zeiss).
PEX1-MO (5’-AA GTG ACC AAC CTG TTC TCC ATC TCT-3’; Gene Tools, Philomath, OR, USA) and Con-MO (5’-AA CTG ACC ATG CTG TTC TCG ATC TCT-3’; Gene Tools) was injected into one-cell stage zebrafish embryos as described in Yeo & Chitnis (2007). Embryos were studied at 25- and 27-hpf.
Mixture of synthetic mRNA of Cas9 (250 ng/μL) and 50 ng/μL sgRNA (5’-GAC AGT CTG CAC TAA CAT ATG G-3’) was injected into fertilized eggs. Whole single embryos at 24-hpf were lysed (100 mM NaOH, 1M Tris-HCl [pH 8.0]). The 1:10 diluted lysates were used for PCR amplification. PCR products were denatured at 95°C for 10 min and ramped from −2°C per second to 85°C, and then reannealed by ramped from −1°C to 25°C per second. Heteroplex PCR products (10 μL) were incubated with 10 U T7E1 enzyme (New England Bio Labs, Ipswich, MA, USA) at 37°C for 40 min. Products from heteroduplex analysis were electrophoresed on 2% agarose gels (Invitrogen).
Adult zebrafish were anesthetized with Tricaine solution (Sigma-Aldrich). The caudal fin was cut off for genomic DNA preparation. The 1:10 diluted lysates were used for PCR amplification. PCR products (10 μL) were incubated with 10 U NdeI enzyme (NEB Labs, Ipswich, MA, USA) at 37°C for 30 min. Products from restriction analysis were electrophoresed on 2% agarose gels (Invitrogen).
RESULTS
The significant sequence difference between Delta and Jagged as Notch ligands gives rise to the question on possible divergent functions of Notch signaling pathway. To investigate Jagged-mediated Notch signaling and identify putative Jagged2-binding proteins, yeast two-hybrid screen was performed using the J2ICD (1106-1254 aa) as baits to screen the zebrafish whole embryos cDNA libraries (Fig. 1A). In the screen, where the reconstitution of the functional transcription factor of Gal4 DNA binding domain-bait fusion protein and prey-Gal4 activation domain fusion protein activates the LacZ reporter gene, 20 positive clones of 12 genes were identified by J2ICD from 60 million colonies. Sequence analysis identified one of positive clones as the N-terminal domain (NTD) of zebrafish peroxin 1, termed PEX1Y2H (Fig. 1B). PEX1 encodes a member of the AAA-ATPase family, a large group of ATPase associated with the PBDs (Reuber et al., 1997). Human PEX1-deficient cells revealed severe defects in import of peroxisomal matrix protein and peroxisomal membrane proteins (Reuber et al., 1997). A single NTD followed by tandom AAA domain is a common feature of organellar membrane-associating AAA-ATPases, suggesting that a possible function of PEX1 as a protein unfoldase in peroximal biogenesis uses its N-terminal putative adaptor-binding domain (Shiozawa et al., 2004). Previous study showed that an E3 ubiquitin ligase interacts with the ICD of a Notch ligand to promote its ubiquitylation and endocytosis (Itoh et al., 2003). Furthermore, Mib promotes the ubiquitylation of the membrane-bound intracellular domain of zebrafish Jagged2 (J2ΔECD), but not that of ICD form in P19 cell (data not shown). Thus, we hypothesized that the NTD of PEX1 may interact with ICD of Jagged2 in neural progenitors located in the ventral spinal cord.

To test this hypothesis, we investigated whether PEX1 and Jagged2 associate in vitro by performing a co-immunoprecipitation assay with various constructs of zebrafish Jagged2 (Fig. 1A). Two constructs, a Myc-tagged Jagged2 variant and Flag-tagged PEX1Y2H, were cotransfected into COS cells. Cell lysates were prepared and subjected to co-immunoprecipitation using anti-Myc monoclonal antibodies at 24 hours post-transfection. When PEX1Y2H was cotransfected with full-length of Jagged2, its immunoreactive band was strongly detected in Western blots by probing with anti-Flag antibodies (Fig. 1C). PEX1Y2H-immunoreactive bands were also detected in the lanes cotransfected with J2ICD or the membrane-bound J2ICD denoted as Myc-J2ΔECD (Fig. 1C). However, the ICD-truncated form of Jagged2 (Myc-J2ΔICD) failed to recruit cotransfected PEX1Y2H (Fig. 1C).
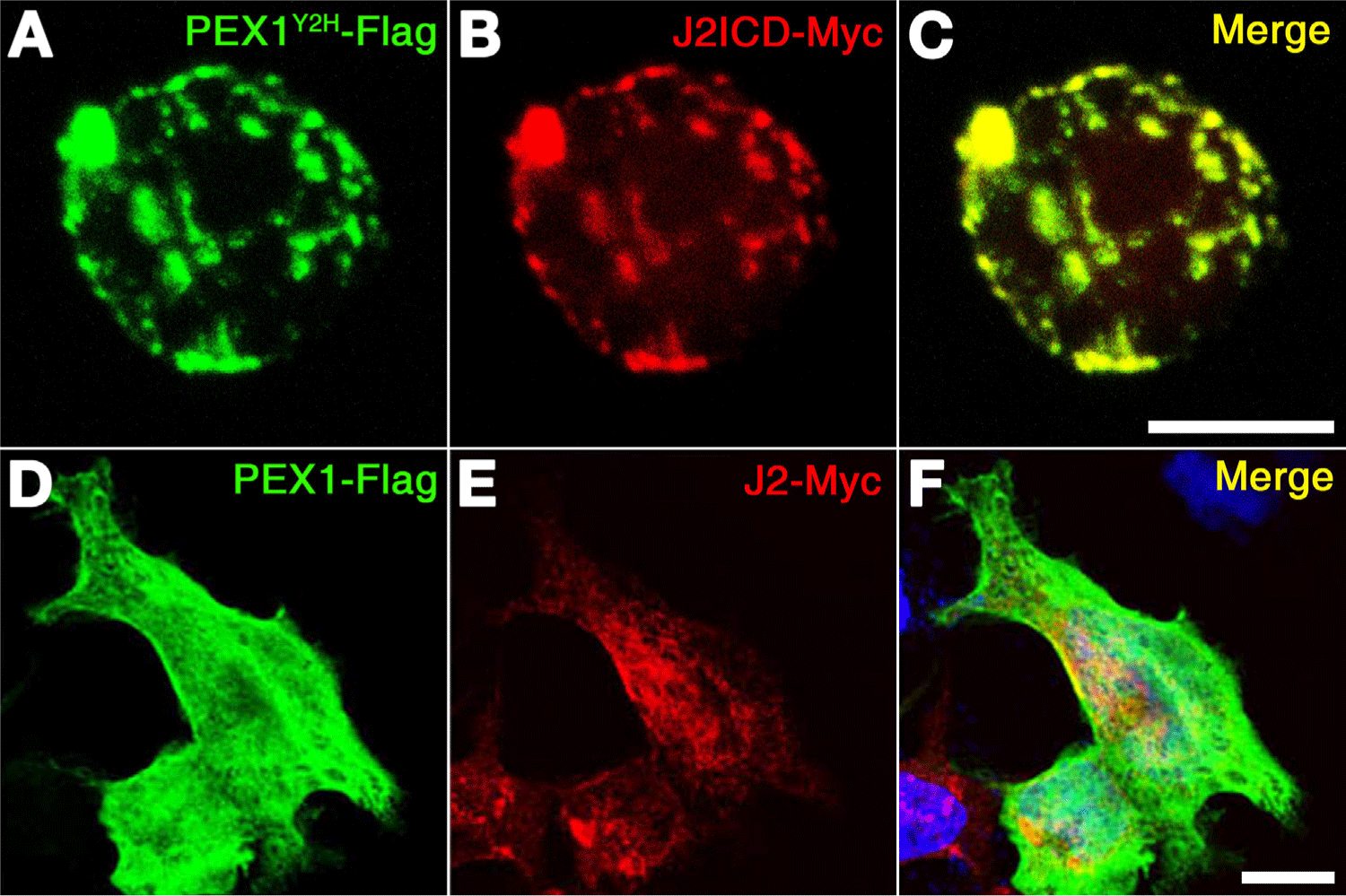
To validate their interaction, colocalization assay was performed by cotransfecting their expression constructs into P19 cells and COS cells (Fig. 2). Since the Jagged-mediated Notch signaling pathway is established in neural progenitors, it seems to be better to use the P19 cell which is epithelial-like morphology isolated from a male mouse with pluripotent embryonic carcinoma (ATCC #CRL-1825). Confocal microscope shows that expression of Flag-tagged PEX1Y2H significantly overlapped with that of Myc-tagged J2ICD in P19 cells, and expression of Flag-tagged PEX1 partially overlapped with that of Myc-tagged J2 in COS cells (Fig. 2). Upon coexpression of PEX1Y2H with J2ICD, PEX1Y2H was found to colocalize with J2ICD in P19 cells to a much higher extent than obtained upon coexpression of Flag-PEX1 with Myc-J2 in Cos cells (Fig. 2). These observations indicate that the NTD of zebrafish PEX1 might interact with zebrafish Jagged2 through its ICD.
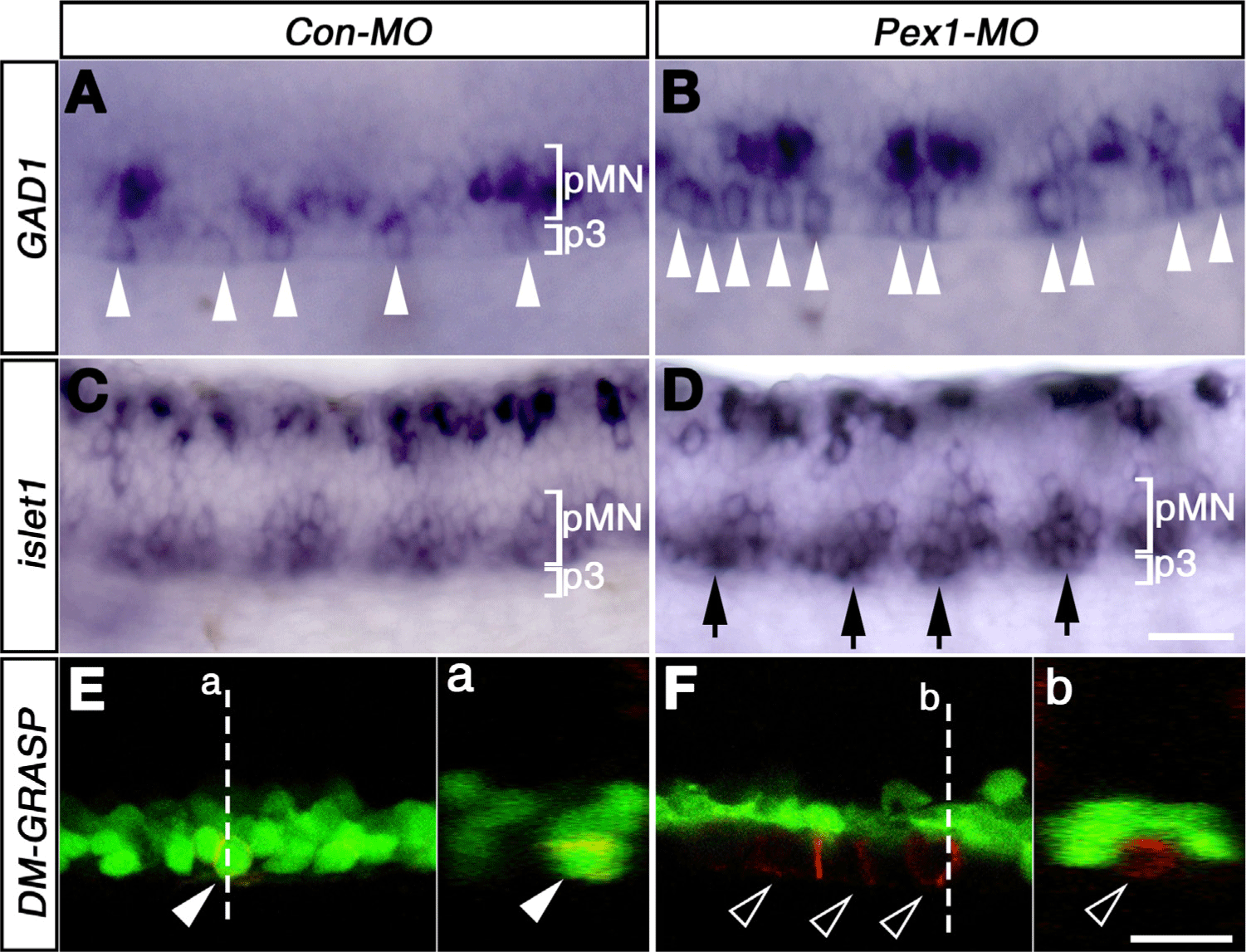
Previous study showed that the number of late-born neurons was limited by Jagged2-mediated Notch signaling (Yeo & Chitnis, 2007). In the ventral spinal cord, motor neurons and KA neurons included KA’ and KA” neurons emerge in a distinct spatio-temporal order from progenitors (Bernhardt et al., 1992). To test whether PEX1 affects the specification of these ventral neurons, a loss-of-function study was attempted by using morpholinos. Morpholino against splicing of PEX1 (PEX1-MO) led to the generation of alternative transcripts, and four bp-mismatched MO (Con-MO) was used as a control (Fig. 3). Morphologically, the embryos injected with MOs developed normally (data not shown). Whole-mount in situ hybridization showed that the number of GABAergic KA” neurons meaningfully increased in the p3 domain of PEX1-MO injected embryos at 25-hpf (~1.4±0.2-fold, mean±SD, p<0.05, n=7) (Fig. 3B). An increase in the number of motor neurons, marked by islet1 expression, was also observed at 25-hpf in the PEX1-MO-injected embryos (Fig. 3D). As the Jagged2 morphants, the some of the ectopic islet-positive cells were located in the most ventral part of the spinal cord (Fig. 3D, Yeo & Chitnis, 2007). At 27-hpf, this remarkable feature was confirmed by staining with zn5 antibody, which labels a cell-surface protein, DM-GRASP, of sMN in the pMN domain (Fig. 3E). EGFP expression in the pMN domain of Tg[olig2:egfp] line provided a criterion for determining ectopic expansion of zn5-positive cells located ventral to the EGFP-expression domain (Fig. 3F). The increase in the number of KA″ cells, together with an increase in that of sMN, indicated that PEX1-mediated signaling limits the number of neurons in the ventral spinal cord.
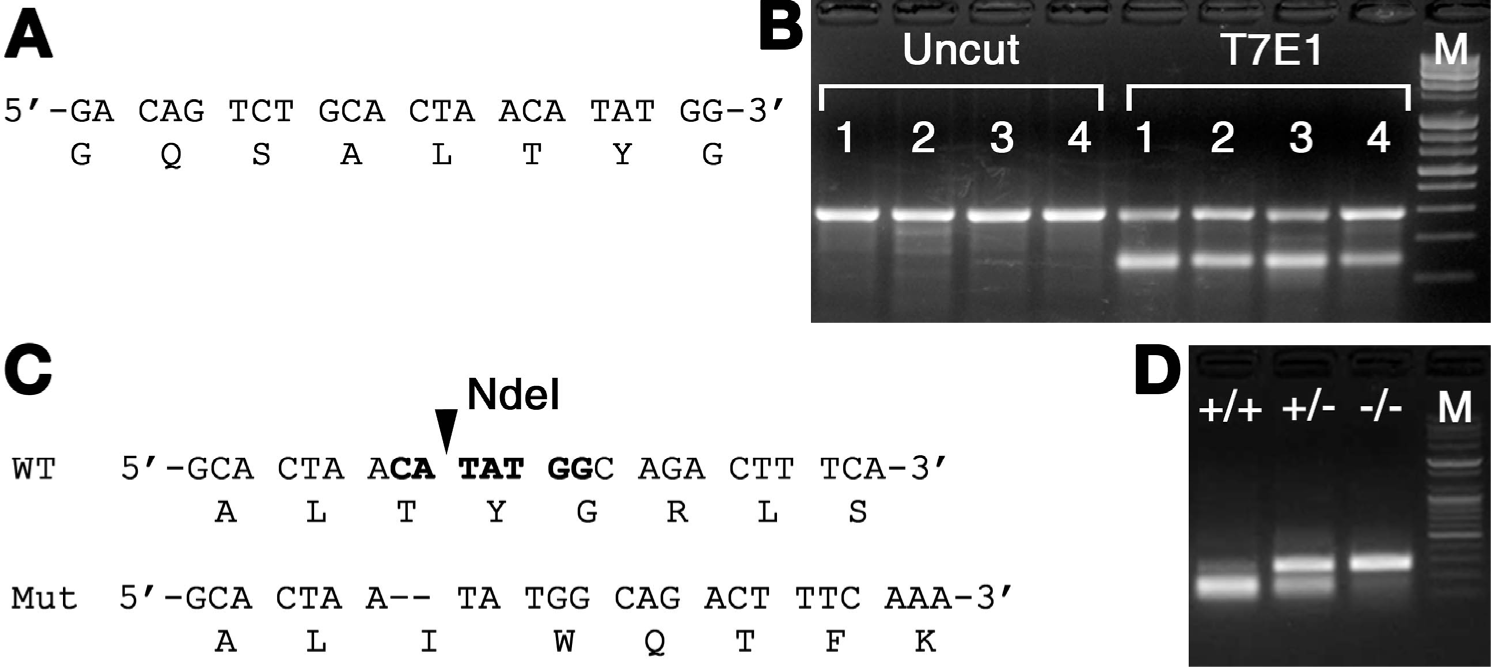
Since these observations need to be supported by establishing a stable zebrafish mutant for PEX1, we designed a guide RNA for CRISPR/Cas9 system (Fig. 4A). The mixtures of synthesized sgRNA and Cas9 mRNA were injected into one cell of fertilized zebrafish embryos. The injected embryos were cultivated at 24-hpf, randomly selected embryos were lysed to investigate the efficiency of the target site mutation generated by sgRNA using T7E1 assay (Fig. 4B). T7E1 assay showed that multiple bands in two out of four embryos. Most individuals showed significant a second major band pattern, indicating DNA heteroduplexes formed and cleaved by T7E1 (Fig. 4B). It was confirmed that insertions and deletions (indels) were constructed through the sgRNA, and PEX1 could be successfully destroyed by the CRISPR/Cas9 system in zebrafish. F1 generation was established by inbreeding selected F0 adults and assessing their genotypes by PCR and sequence analysis to select individuals (Fig. 4C). A total of 24 F1 individuals were screened and analyzed. Several mutation patterns were observed, three of which affected the reading frame, and the obtained sequence pattern resulted in a deletion of two base pairs (Fig. 4C). In the F2 generation, three genotypes were obtained: wild-type (PEX1+/+), heterozygotes (PEX1+/−), and homozygotes with 2-bp deletion mutation (PEX1−/−). Detection of the three genotypes was performed using a polymerase chain reaction-restriction fragment length polymorphism ass (PCR-RFLP) (Fig. 4D). The amplified DNA fragments from wild-type were digested by NdeI restriction enzyme, but not from the mutant with a 2-bp deletion (Fig. 4D). RFLP analysis confirmed PEX1 homozygous mutation and isolated female PEX1 mutants.
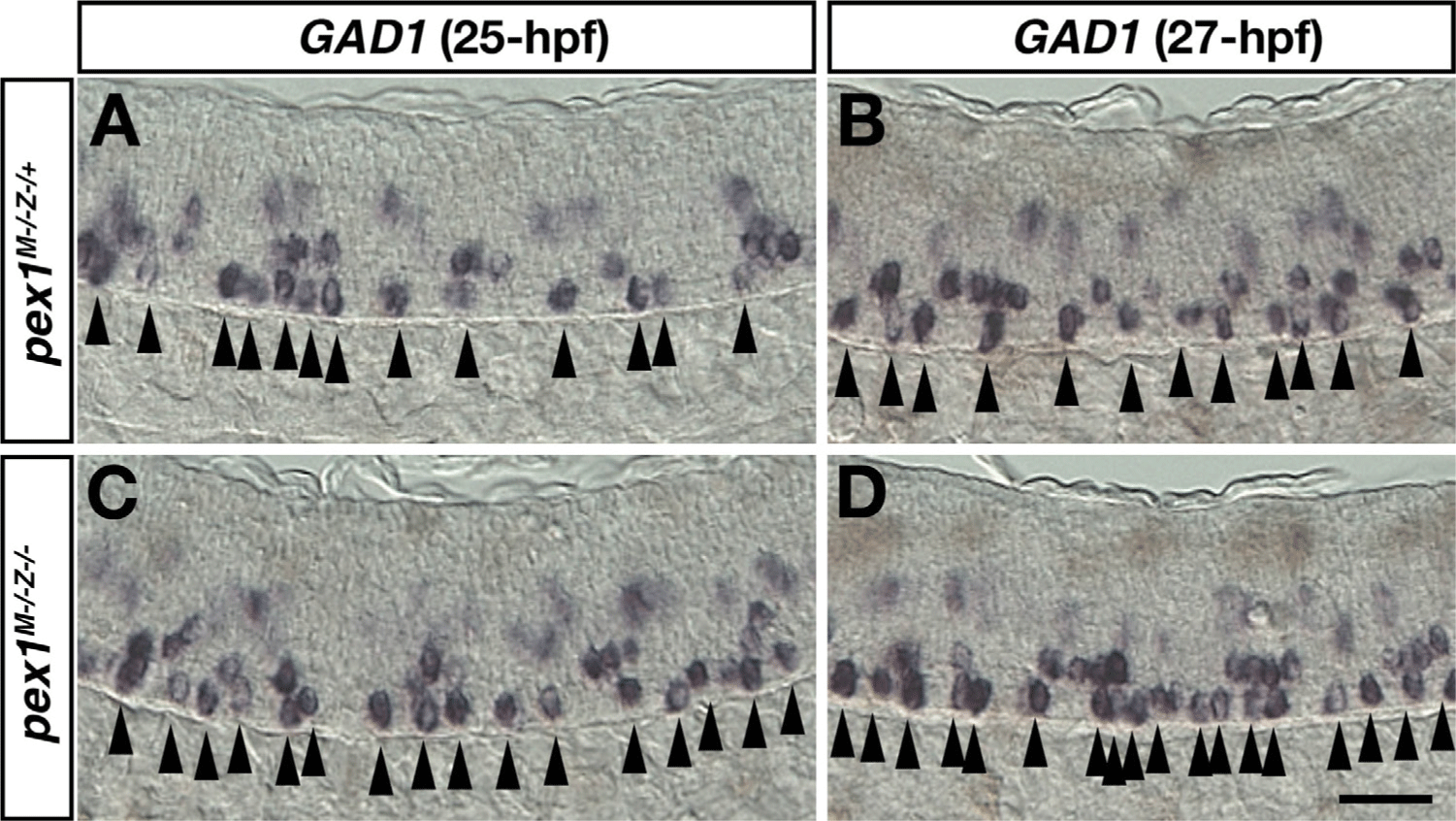
To confirm whether PEX1 mutation affects the specification of KA” neurons, whole-mount in situ hybridization was performed using the GAD1 riboprobe (Fig. 5). Fertilized eggs of homozygous PEX1 mutant were obtained by mating with homozygous PEX1 female mutants and heterozygous male mutants. The number of GABAergic KA” neurons slightly increased in the p3 domain of maternal-zygotic PEX1 mutant at 25-hpf (~1.2±0.01-fold, mean±SD, p<0.05, n=5), although the number of KA” cells is variable (Fig. 5C). At 27-hpf, an increase in the number of KA” neurons was also observed in another batch of maternal-zygotic PEX1 mutant (Fig. 5D, ~1.5±0.05-fold, p<0.01, n=5). The increase in the number of KA″ cells was consistently observed in transient knock-down PEX1 morphant and permanent knock-out PEX1 mutant, suggesting that PEX1-mediated signaling limits the number of neurons differentiated from p3 progenitors.
DISCUSSION
The development of late-born neurons, including sMN and some GABAergic neurons in the ventral spinal cord, requires specific mechanisms that maintain neural progenitors in an undifferentiated state until the appropriate time comes. When released from these mechanisms, neural progenitors could differentiate and assume the appropriate fate (Anderson, 2001). Previous study showed that Jagged-mediated Notch signaling is the key component of these mechanisms and plays an important role in maintaining pluripotency of neural progenitors in the ventral spinal cord until they assume a specific fate (Yeo & Chitnis, 2007). In the present study, our observation provides the possibility that PEX1 could be a novel component of Jagged2-mediated signaling pathway, suggesting that PEX1 might play an auxiliary role in signaling cells to activate Notch signaling pathway in neighboring cells by Jagged2.
Throughout yeast-two hybrid screening using zebrafish Jagged2 ICD as a bait, NTD of zebrafish PEX1 was isolated. This study showed physical interaction between zebrafish PEX1 and zebrafish Jagged2 by co-immunoprecipitation, and examined the role of PEX1 in neurogenesis by using its knockout mutant. PEX1 is a component of a protein transport system comprising more than 30 proteins which is involved in biogenesis and maintenance of the peroxisome (Brown & Baker, 2003). Peroxisomal protein transport requires ATP. Of the more than 30 peroxins, only two AAA-ATPases, human PEX1 (hPEX1) and human PEX6 (hPEX6), have been identified (Reuber et al., 1997; Geisbrecht et al., 1998; Shiozawa et al., 2004). Defects in the ATPase activity of hPEX1 or hPEX6 lead to impaired peroxisome biogenesis (Geisbrecht et al., 1998). Although the two AAA-ATPase domains of hPEX1 and hPEX6 display a high sequence identity, the N-terminal sequences of hPEX1 and hPEX6 are quite different (Shiozawa et al., 2004). The crystal structure of PEX1 NTD reveal a similarity to that of the N-domain of VCP and NSF (Shiozawa et al., 2004). NSF is responsible for membrane fusion in exocytosis (Hay & Scheller, 1997), and VCP are involved in ER-associated protein degradation as well as remodeling of the Golgi (Kondo et al., 1997). The common features these proteins is components of vesicle transport. This domain architecture is known as a supradomain as found in putative binding domain to Jagged2 ICD. These observations are reminiscent of a previous study in which Notch activation is followed by endocytosis of ubiquitinated Delta by Mib (Itoh et al., 2003). The ICD of zebrafish Jagged2 was also ubiquitinated by Mib (unpublished data). Our data, together with these observations suggest a model that Notch activation might be followed by PEX1-mediated endocytosis of ubiquitinated Jagged2 by Mib and trans-endocytosis of the Notch ECD by the signal-sending cell, such as neural progenitors in the ventral spinal cord.
When KA” neurons are born from the neural progenitors in the most ventral p3 domain of spinal cord, it is required that Jagged2 expressed in pMN domain activates Notch signaling pathway in p3 progenitors (Yeo & Chitnis, 2007). In wild-type zebrafish embryos, the neural progenitors in the p3 domain divide into two between 9-hpf and 16-hpf; one becomes KA” neuron, whereas the other should remain as a progenitor for next stage of neurogenesis. Therefore, it seems reasonable that the number of KA” neurons in Jagged2 morphants was increased approximately 1.9±0.2-fold (Yeo & Chitnis, 2007). In addition, the number of KA” neurons increased dramatically by more than 4-fold due to the disruption of Notch signaling caused by overexpression of Mib:EGFP (Ryu et al., 2015). Although PEX1 morphants showed phenotypical similarity to Jagged2 morphants, the number of KA” neurons in PEX1 morphants was increased approximately 1.4±0.2-fold, and that in PEX1 deletion mutants was only increased approximately 1.2+0.01-fold. These observations imply that Notch signaling pathway may be partially impaired by knockdown of PEX1, suggesting a possibility that there might be functional redundancy among transporter-related genes, such as PEX6, VCP and NSP (Shiozawa et al., 2004). Conservation of structural motifs reflects the conserved functions of peroxins during evolution and development, but individual subtypes of PEX protein family likely contribute in unique ways to building the overall peroxisomal architecture. Or these observations suggest other possibility that a permissive factor requires for activation of Jagged2 with PEX1 to fully active Notch signaling pathway. Previous study showed that generation of KA” neurons required the coordination of retinoic acid and Mib-mediated pathway (Ryu et al., 2015). The relationship between PEX1 and Mib could be investigated by generating PEX1 mutant in a Tg[hsp70-Mib:EGFP] transgenic background. By treating PEX1−/−::Tg[hsp70-Mib:EGFP] zebrafish with retinoic acid at several stages, the formation of secondary motor neurons and KA” neurons could be observed. In this respect, PEX1 mutant may be a useful tool for generating combinatorial genetic modified zebrafish, and the increase in the number of KA” neurons may also be a useful criterion for evaluating Jagged2-mediated Notch signaling pathway in the ventral spinal cord.
