INTRODUCTION
The testes produce spermatozoa via the spermatogenesis. The spermatogenesis is the process by which haploid spermatozoa are generated from spermatogonia cells, which are stem cells, in the seminiferous tubules of the testes. The spermatozoa travel very long way in male reproductive tract. On the course of the journey they meet the substances secreted from male reproductive organs and obtain capacity to fertilize the eggs subsided in the fallopian tube following ovulation in female reproductive tract.
The process is common in mammals. Like rat and mouse investigated widely, Syrian hamsters living in temperate zone follow the germ cell-forming procedure mentioned above. But the reproductive activities of Syrian hamsters are arrested in the winter climate (Choi & Lee, 2012). In fact Syrian hamsters show energetic breeding in summer climate but infertile during winter season, which represents seasonal breeding (Choi, 2013a). The cyclic changes of reproductive function have been well established to be regulated by photoperiod (Stetson & Watson-Whitmyre, 1984; Stetson & Watson-Whitmyre, 1986). Long photoperiod (LP; equal to or greater than 12.5 hours of lightings in a day) sustains large testes but short photoperiod (SP; equal to or less than 12 hours of lightings in a day) reduces visibly the mass of testes. Thus the spermatogenesis is vigorously developed in LP and ceased completely in SP (Choi & Han, 2010; Choi & Lee, 2012). The phenomenon renders the animals to spare the energy, to use it efficiently, and to conserve the species unimpaired. The natural fluctuating changes of seasonal reproductive function in the hamsters can be recapitulated in the artificial lighting regime. When the reproductively mature male hamsters are transferred to SP, they lose generative activities in near two full months, showing reduced testes in size and no functional spermatozoa in testes (Reiter, 1980). If the length of lighting in a day is set to LP imitating summer season, the generative functions are energetically resumed, doubtlessly promoted, and maintained afterward (Stetson & Watson-Whitmyre, 1986).
Spermatozoa formed in the testis undergo a post-testicular maturation process through the male reproductive track. In the process they are exposed to diverse substances secreting from the Sertoli cells, epididymal and vas deferens epithelia, and seminal vesicle and prostate cells. The surface membrane of the spermatozoa experiences various modification events, undergoing both protection from microorganisms and reconstruction of functional spermatozoa (Cohen et al., 2001; Dacheux et al., 2003; Jalkanen et al., 2006; Nixon et al., 2006).
Epididymal protease inhibitor (EPPIN), which is synthesized and secreted from the Sertoli cells of the testes and epididymal epithelial cells, is one of the sperm surface proteins. It is encoded by the WFDC genes, which is located in chromosome 20, in human (Richardson et al., 2001; Clauss et al., 2002). EPPIN was also identified in mouse (Sivashanmugam et al., 2003) and rat (Bian et al., 2009). It protects the spermatozoa from the microbial attaks (Yenugu et al., 2004), provides a site for the seminal plasma protein semenogelin (SEMG) to bind (Wang et al., 2005), and involved in the degradation of the EPPIN-SEMG complex by prostate specific antigen (PSA) enzyme (O’Rand et al., 2006). EPPIN on the surface of spermatozoa has been described to bind to SEMG, clusterin, and lactotransferrin (Paasch et al., 2011). It intervenes between the surface membrane of spermatozoa and various substances (Zhang et al., 2013). The ejaculated spermatozoa are coated with SEMG and undergo a temporary retardation of forward motility. The progressing motility is resumed at the time wheh the PSA works for the degradation of the complex. Persistent SEMG on the surface of spermatozoa results in seminal hyperviscosity and infertility (Esfandiari et al., 2008; Emami et al., 2009; Du Plessis et al., 2013).
When the EPPIN binds to anti-EPPIN antibodies or SEMG, the progressive motility is reduced steeply not to be able to reach to the ovulated egg (O’Rand et al., 2004; O’Rand et al., 2009; Mitra et al., 2010). This situation results in infertility, inciting a development of male contraceptive agent. A small compound to be capable of diminishing the speramatozoa motility was explored as a potential non-hormonal male contraceptive (O’Rand et al., 2018). Those researches raise a question whether the reproductive capability is related to the expression of EPPIN gene in reproductive organs.
The goal of the present work was to identify the EPPIN gene, to compare to genes reported previously, and to associate with reproductive capability in the male Syrian hamsters.
MATERIALS AND METHODS
Male Syrian hamsters (Mesocricatus auratus) were used to identify the EPPIN gene. They were housed in plastic cages under LP conditions of light and dark (lights of 14 h:darkness of 10 h) with an ambient temperature of 22±1℃. Reproductive activities of these hamsters in 8 weeks after birth are always active in the photoperiod. Spermatozoa were confirmed in both the testis and the epididymis by the microscopic examination. The animals were fed with standard laboratory mouse chow and tap water ad libitum. Some of the reproductively mature Syrian hamsters were transferred to SP (lights of 10 h:darkness of 14 h) and kept for 8 weeks during which their reproductive activity is entirely degenerated. Various tissues including the reproductive organs were extracted and immediately subjected to the reverse transcription polymerase chain reaction (RT-PCR). The condition of management of animals was approved by the Yongin University Institutional Animal Care and Use Committee (YUIACUC-2017-02).
The expression of EPPIN gene was examined in the testis and epididymis of the Syrian hamsters near 1 week intervals for 8 weeks after birth. The testes and epididymis tissues of the animals at 3 days after birth were pooled because they are very small. Afterward the tissues of testes and epididymis from an animal were available and used to perform RT-PCR of the EPPIN gene.
The male Syrian hamsters were sacrificed by decapitation. Immediately the testes were excised and the suitable parts of them were immersed in the physiological saline. The tissues were cut many times with sterile scissors without delay. Following 1 min at room temperature, 1 mL of supernatant was transferred into microcentrifuge tube. The tube was spun at 14,000 rpm for 1 min (CentrisartⓇA-14C, Sartorius, Germany). The supernatant was removed and 1 mL of saline was added to the tube. The pellet in the tube was dispersed by sucking and releasing of pipette. One drop of the suspension was fallen onto the slide glass and smeared uniformly. Then the slide glass was dried completely and absolute methanol was fully applied. After the entire dryness of the slide, specific staining solution (hematoxylin) was employed and dispersed. Ten minutes later, the extra staining solution was cleanly washed with flowing tap water. Then the presence of spermatozoa was observed by light microscope (Leica DM500, Leica Microsystems, Switzerland).
The EPPIN gene of Syrian hamster has not been identified up to now. Thus primer sequences for EPPIN gene of the animal were chosen from the mRNA predicted in the NCBI Reference Sequence (XM_013124401.1). The primers selected were 5’-ggagatgccccc/agattcaga-3’ for forward direction and 5’-tggaagttgttattgtttcc-3’ for reverse direction. The anticipated size was 262 bp. The primers have high homology in comparison to the sequences of mouse and rat. Glyceraldehydes-3-phosphate dehydrogenase (GAPDH) PCR was used as reference standard for RT-PCRs in the present study. The primers of GAPDH were 5’-aaatga ccccttcattgacc-3’ for forward and 5’-ccttccacaatgccaaagtt-3’ for reverse. The anticipated size was 420 bp. Sequence analyses were done by a commercial sequencing service company (Bioneer, Korea).
Total RNAs were isolated from tissue samples using TRIzolⓇ Reagent (Invitrogen, USA) according to the manufacturer’s protocol. That is, the small pieces of tissues (50–100 mg) were excised and subjected to sonicate with 1 mL of TRIzolⓇ Reagent (VCX130, Vibra CellTM, Sonics & Materials Inc., USA). The samples were transferred to new microcentrifuge tubes and spun for 5 min at 12,000 rpm at 4℃. The supernatant was moved into the new tubes and left for 5 min of incubation, allowing to permit complete dissociation of the nucleoprotein complex. 0.2 mL of chloroform was added and capped firmly the tubes. Following the incubation of 2–3 min, the tubes were spun for 15 min at 12,000 rpm at 4℃. The upper aqueous phase was transferred to the new tubes. A half mL of isopropanol was added and incubated for 10 min. Then the tubes were spun for 10 min 12,000 rpm at 4℃. The supernatant was discarded and the pellets were resuspended in 1 mL of 75% ethyl alcohol. After agitation, the samples were spun for 5 min at 7,500 rpm at 4℃. The supernatant was eliminated and the pellets were allowed to dry for at least 5 min. The pellet was solubilized with 20–50 μL of RNase-free water. Quantitation of the RNA was measured by the absorbance at 260 nm that provides total nucleic acid content and 280 nm that determines purity of the RNA.
The extracted RNAs were used in RT-PCR reactions carried out with Maxime(tm) RT PreMix and AccuPower PCR Premix (Bioneer, Korea) according to the manufacturer’s instructions. Reverse transcription was primarily carried out to create complementary DNAs (cDNAs) representing cell-specific RNA populations. The proper amount (1 pg-1 μg) of tRNA was transferred to clean microcentrifuge tubes and mixed with the following materials: DEPC-treated water, reverse transcription reaction buffer, oligo (dT) 20 primer, dNTPS (dATP, dTTP, dCTP, dGTP), reverse transcriptase, and RNase inhibitor. The tubes were gently agitated and incubated at 42℃ for 60–90 min. In order to inactivate the reverse transcriptase the tubes were heated to 85℃ for 5 min. The cDNA products transcribed were stored at -20℃.
PCR was performed with the cDNA diluted with TE buffer (10 mM Tris (pH 8.0), 0.1 mM EDTA). The microcentrifuge tubes with template cDNA (typically 10 ng) were mixed with water, 10x PCR Buffer, dNTP Mix, primers (forward and reverse), Taq DNA Polymerase, and 25 mM MgCl2. The tubes were stirred gently by vortexing and spun briefly to collect all components to the bottom of the tubes. The cycles of PCR were 40 with repeating the following in the order: denaturing temperature of 94℃ for 20 seconds, annealing temperature of 55℃ for 30 seconds, and extension temperature of 72℃ for 1 min. The final extension was performed at 72℃ for 5 min and then cooled down to 4℃.
The reaction products were analyzed by gel electrophoresis in 1.5% agarose gel (100 V, 60 min) and visualized by ethidium bromide staining. The bands were identified using the image analysis system (Chemi Doc XRS, Bio-Rad, USA).
The PCR products were purified through the agarose gel electrophoresis according to the manufacturer (AccuPrepⓇPCR/Gel Purification Kit, Bioneer Corporation, Korea). The PCR products were subjected to the electrophoresis and stained with ethidium bromide. The visualized gel bands were cut out using blade. The gel slices were mixed with 3 volumes of FB buffer. The tubes were incubated at 50℃ for 10 min with mixing by inverting every 2–3 min. One volume of absolute isopropanol was added and mixed immediately by inverting. The mixture was transferred to a binding column in a 2 mL collection tube. The lid was closed and spun at 14,000 rpm for 1 min. The binding column was reassembled with collection tube after removing the flow-through fluid. 500 μL of W2 buffer was added and spun at 14,000 rpm for 1 min. The binding column was reassembled like above. Then the step with W2 buffer was repeated once more and spun at 14,000 rpm for 1 min. The binding column tube was transferred to a new 1.5 mL tube for elution. 30 μL of EA buffer was added carefully onto the binding column tube and waited for at least 1 min at room temperature. Finally the new tube was spun at 14,000 rpm for 1 min. The eluant was sent to Bioneer (Korea) to analyze the sequence of EPPIN gene.
RESULTS
The male Syrian hamsters housed in LP for 8 weeks showed large testes that was 1.6 g of average of each testis, which represents full spermatogenesis. The expression of EPPIN gene was primarily detected in testis of male Syrian hamsters (Fig. 1, left). The size of the gene was 262 bp as expected. In order to confirm the tissue-specific expression of EPPIN gene, various organs of both male and female Syrian hamsters, including heart, lung, liver, stomach, spleen, adrenal, kidney, skeletal muscle, testis, epididymis, and seminal vesicle were examined. Among many other organs, solely both testis and epididymis of males showed the expression of EPPIN (Fig. 1, middle). Females did not show the gene at all in any tissues examined (Fig. 1, right). The size of the GAPDH used as reference standard for RT-PCRs was 420 bp as expected.
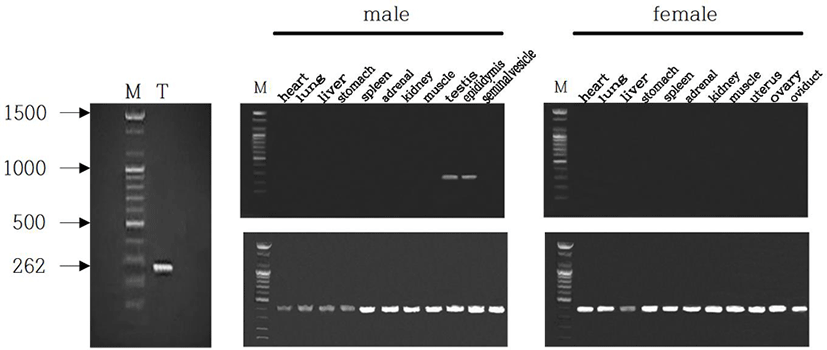
The mRNA of EPPIN gene of Syrian hamster that identified in this investigation was compared to other EPPIN genes of chinese hamster, rat, mouse, and human reported in the range of sequence detected (Fig. 2). The sequence of EPPIN of the Syrian hamster was exactly identical to the sequence announced as predicted in NCBI reference sequence XM_013124401.1 (LOC101834060) and had homology of 96.9% to the sequence of Chinese hamster (Cricetulus griseus) EPPIN-like mRNA predicted (LOC10 0766120). The sequence of EPPIN of rat was also determined by using the same primers at the same time. The sequence of EPPIN gene identified in the Syrian hamster had homology of 91.2% to that of rat and 88.5% homology to that of mouse. In comparison to the human EPPIN gene, it showed only 75.2% homology.

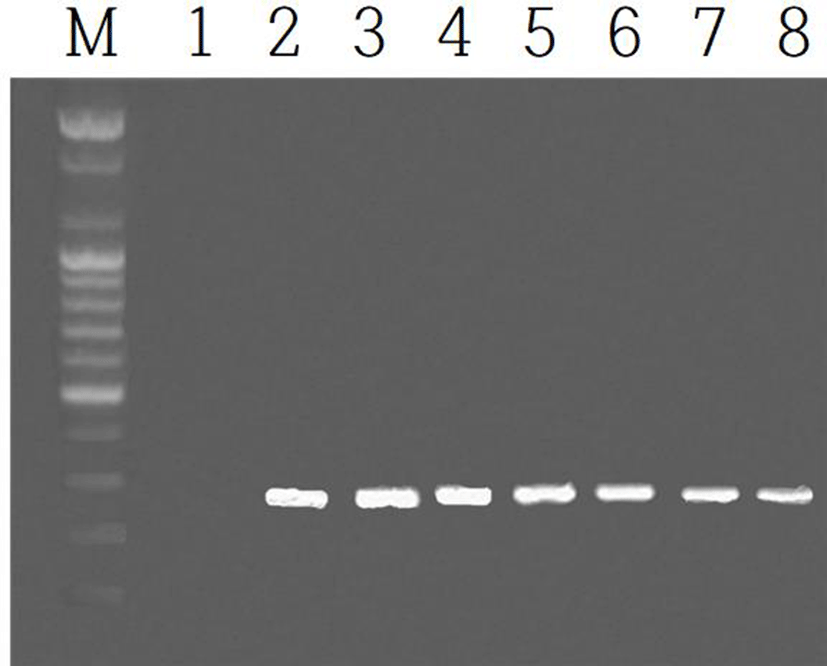
The expression of EPPIN gene was examined in the testis and epididymis during the course of development after birth. The expressions of the gene in the testes were shown in Fig. 3. The identical results of EPPIN gene were observed in the epididymis. The EPPIN mRNA was not detected in the age as early as postnatal 3 days (marked as 1 in Fig. 3). The gene began to observe from the 2nd week after birth (indicated by 2 in Fig. 3) and continued to the age of 8 weeks at which the animals begin to breed energetically.
In order to inspect the relationship of EPPIN and fertility capability in male Syrian hamsters, the expression of EPPIN gene was investigated in the two functionally different testes: reproductively energetic testes and completely regressed testes of the Syrian hamsters maintained in LP and SP, respectively. The Syrian hamsters housed in LP had large testes of 1.6±0.27 g of mean weights of testes and the animals maintained in SP for 8 weeks showed very small testes of 0.2±0.02 g of mean weights of testes (Fig. 4a and 4b). The LP animals had abundant spermatozoa in testis but the SP animals showed no spermatozoa at all in the organ tissue (Fig. 4c).
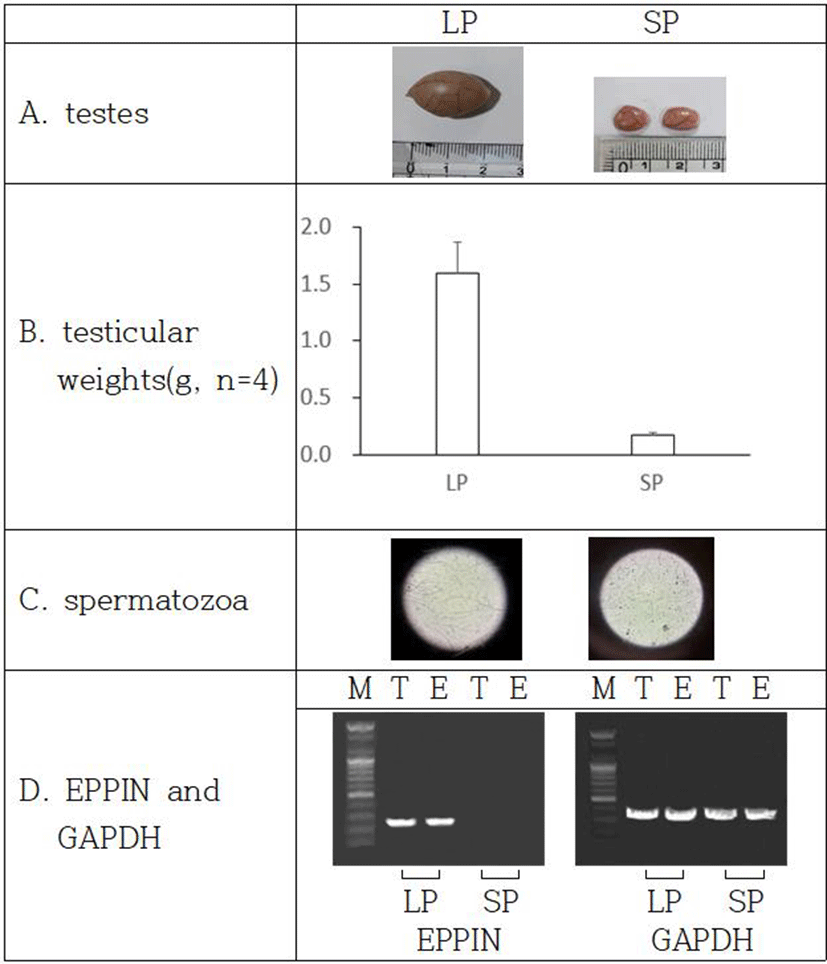
The expression of EPPIN gene was witnessed in only testes and epididymis of the reproductively active Syrian hamsters kept in LP (Fig. 4d). The gene was not detected in the SP-induced small testes, which is an indicative of cessation of spermatogenesis.
DISCUSSION
The present results represent the association of the EPPIN gene with the fertility capability in male Syrian hamsters. Only reproductively active Syrian hamsters expressed EPPIN genes in testis and epididymis. Females did not demonstrate any EPPIN gene at all. In the sexually regressed testis of SP animals, EPPIN mRNA was not detected in testis and epididymis. This is the first report that the EPPIN gene is directly related to the fertility ability in these animals.
As well-established, the Syrian hamsters maintained in LP show large mass of testes, indicating energetic and functional spermatogenesis (Stetson & Watson-Whitmyre, 1984; Stetson & Watson-Whitmyre, 1986). As a seasonal breeding animal, the Syrian hamsters display completely regressed testes in winter climate of natural environment or in 8 weeks of SP in laboratory facility. The degenerated testes are evidenced by shortened diameter of more than one half of seminiferous tubule (nearly one eighth in volume), indicating the absence of germ cells experiencing meiosis (spermatozoa and spermatocytes) in the histological examination (Choi, 2013b). The EPPIN gene was only expressed in testes and epididymis of the sexually active male Syrian hamster. These results are consistent with the consequences reported from human, mouse, and rat (Richardson et al., 2001; Sivashanmugam et al., 2003; Bian et al., 2009). Female Syrian hamsters showed no expression of the EPPIN gene as previously reported in other animals (Bian et al., 2009). The integration of the outcome signifies that the gene is male-specific gene, especially expressing only in designated reproductive organs testis and epididymis.
When the EPPIN gene identified in male Syrian hamster was compared to the genes of other species (Fig. 2), it had very high proportion of homology, impling the same function mentioned in other reports (Richardson et al., 2001; Shivashanmugam et al., 2003). The sequence of EPPIN gene of the Syrian hamster had homology of nearly 90% to that of rodents (96.9% to Chinese hamster, 91.2% to rat, and 88.5% to mouse). It showed 75.2% homology to the human EPPIN gene. The high similarity denotes that the gene has been conserved from the ancestry (Silva et al., 2013).
The postnatal expression of EPPIN gene was detected in both organs testes and epididymis in the age of postnatal 2nd week. Immediate examination at 3 days after birth did not show the EPPIN mRNA. The expression of EPPIN gene was continued to the age of 8 weeks at which the animals begin to breed energetically, which is similar results reported previously (Bian et al., 2009). The consequence signifies that the sexual capability during the course of growth including puberty is being earlier prepared before the spermatozoa are produced in the male gonads.
The EPPIN protein provides a base to be able to reduce human sperm motility, preventing the spermatozoa from fertilizing the egg (O’Rand et al., 2011; O’Rand et al., 2016; O’Rand et al., 2018). The variants to the gene has also been reported to associate with male infertility (Ding et al., 2010). Moreover, The antibody to the EPPIN protein also raise a possibility as a contraceptive by inhibiting the action of the subsequent protein in the pathway of the spermatozoa to gain fertilization capability (O’rand et al., 2004). Thus the gene product presents a notable site to develop an anticonceptive agent.
In the male Syrian hamsters whose sexual activity was completely arrested by SP, the EPPIN gene was not expressed in both testes and epididymis. The distinguished regression was assured by the reduced size, lowered weights of testes, and the absence of spermatozoa (Choi & Lee, 2012). Accordingly, the results demonstrate that the EPPIN gene could be a possible biomarker because the gene is exclusively expressed in the reproductively energetic animals. It has not been known when the expression of the EPPIN gene is inhibited in the course of SP environment. A factor (or factors) that prevent the EPPIN gene from expressing in the reproductive organs also has not been known at this time. The inquiry remains to be investigated.
On the other hand, the function of EPPIN protein mentioned above raises a reasonable possibility that a natural product would exert as a contraceptive agent if it binds to the EPPIN protein on the surface of spermatozoa. EPPIN protein can be bound by another protein SEMG, which provide a reaction site for PSA, resulting in the increase of spermatozoa motility. By the binding of a natural product to the EPPIN, if SEMG can not bind to the EPPIN and PSA is unable to recognize the EPPIN-SEMG complex, the capability of the spermatozoa to fertilize the egg will be sharply reduced. The incapability will be appeared by increase of viscosity, reduction of motility, and inhibition of action of PSA.
In conclusion, male Syrian hamsters expressed EPPIN genes in both testis and epididymis and females did not show any EPPIN genes at all, meaning the male-specific gene. In the reproductively regressed testis of SP animals, EPPIN mRNA was not detected in testis and epididymis. The results suggest that the expression of the EPPIN gene is associated with fertility capability in male Syrian hamsters. It is a future work that when and how the expression of the EPPN gene is controlled in the males housed in SP.
