INTRODUCTION
The marine medaka, Oryzias dancena, is a euryhaline teleost that mainly inhabits river mouths and estuaries, occurring in brackish water or freshwater around Bengal Bay and the Malay Peninsula (Roberts, 1998). This species is a bony fish with high tolerance of salinity because of its salinity adaptation mechanisms, and is widely used as an experimental fish. Inoue and Takei (2003) have used marine medaka as a molecular biomarker and as an experimental fish in studies investigating adaptations to seawater. In addition, its viability under conditions of maximum tolerable salinity has been measured, and incubation party-fry in times was assessed in variety salinity its ability to adapt (Nam et al., 2010). Kim et al. (2009a) studied the early gonadogenesis and sexual differentiation of the marine medaka, and found that its gonadal differentiation is the typical type of differentiated gonochorism.
Until now, this species was selected by the Institute of Marine Living Modified Organisms (iMLMO), Pukyung National University, Korea, for a living modified organism evaluation project. Consistent with this purpose, detailed information on its biology, especially its early gonadogenesis, sexual differentiation, early ontogenesis, embryogenesis and exceptional capacity for hyperosmoregulation and hypoosmoregulation, and is becoming available (Kim et al., 2009a, 2009b). In addition, Kim et al. (2009a) suggested that this species has a short interval between generations with spawning possible only 60 days after hatching. Park et al. (2011) studied the effects of clove oil and lidocaine-HCl on it, and the results of their study have contributed to the safe laboratory handling of this species, which is used extensively in research. These studies have demonstrated that the marine medaka has the necessary attributes of an experimental animal (Kim et al., 2009a, 2009b; Nam et al., 2010; Park et al., 2011).
Triploidization is a technique used to generate sterile aquatic animals by exploiting the mispairing of their three homologous chromosomes during meiosis I (Don & Avtalion, 1986). This technique also enhances the productivity of several fish species because it is assumed to increase yield by sequestering the energy usually required for gonadal development to somatic growth (Tave, 1993). More importantly, it generates fish that cannot breed and consequently cannot contribute to the local gene pool if they escape from culture. By conferring sterility on exotic fish for a limited purpose, triploidy is an effective method of reducing or eliminating the environmental risks imposed by genetically modified organisms (Dunham & Devlin, 1999).
Triploid animals generally have similar, if not identical, morphological and meristic characteristics to those of their diploid counterparts (Bonar et al., 1988). However, several morphological differences and abnormalities have been associated with triploidy in fish. A variety of deformities have been reported in the triploid pejerrey, Odontesthes bonariensis (Strüssmann & Takashima, 1990), but it is unclear whether the fish examined were in fact triploid or aneuploid (i.e., having a chromosome number other than a complete multiple of the haploid genome). Flajshans et al. (1993) described differences in the pelvic fin shapes and lengths of triploid and diploid tench, Tinca tinca, and Tave (1993) observed facial deformities in triploid bighead carp, Hypophthalmichthys nobilis, and grass carp, Ctenopharyngodon idella. Probably the best and most frequently described example of a gross morphological difference in a triploid fish is the development of lower jaw deformities in the triploid Atlantic salmon, Salmo salar (Lee & King, 1994; McGeachy et al., 1996). Although conclusive data are lacking, this deformity may be linked to its rapid growth rate in seawater (Lee & King, 1994).
Morphological differences between species or populations are usually understood and compared in terms of general figures or specific anatomical shapes (Straüss & Bond, 1990). The morphometric characteristics of fish, unlike meristic, countable characteristics, can be measured in millimeters. Although our understanding of the morphometric characteristics of fish is limited because they can be so readily modified by the environment, the general figure of a fish is largely determined by genetic factors (Park et al., 2004). The morphometric characteristics of fish are used in three major ways: to distinguish sexes and species and to identify confusing species, such as crossbred hybrids; to study character modifications in groups and species; and to identify and classify biotypic linkages (Park et al., 2004).
Both truss and classical dimensions are used to describe the fish body shape (Straüss & Bookstein, 1982). Truss dimensions consist of a systematically arranged set of distances that are measured among a set of preselected anatomic landmarks, which are points identified on the basis of local morphological features and are chosen to divide the body into functional units (Straüss & Bond, 1990). Truss dimensions, which include components of body depth and length along the longitudinal axis, have theoretical advantages over classical morphometric characteristics in discriminating groups (Park et al., 2001a). In this study, we examined the possible morphometric characteristics of the marine medaka, in terms of the truss and classical dimensions, that might allow diploid and triploid fish to be distinguished.
MATERIALS AND METHODS
The experimental group of diploid marine medaka, Oryzias dancena, used in this study was reared according to the methods of Park et al. (2011). On 24 September 2015, 100 fish were quarantined into male and female categories and cultured in 100 L glass aquariums for three days. The sex ratio of males:females was 60:40. The culture water was dechlorinated and 30% of the water in the aquariums was changed every day. Artemia salina collected from a culture aquarium were provided to the fish every day as a food source. Fish with a standard length of > 25 mm were used in this experiment and 35 male and 15 female marine medaka were placed in each of two aquariums, and 1,000 fertilized eggs were collected immediately by net. The fertilized eggs of the diploid experimental group (n=500) were reared in a 100 L glass aquarium.
The fertilized eggs of the triploid experimental group (n=500) were left to fertilize for 5 min, and were subjected to cold-shock treatment (4℃) for 60 min to prevent the expulsion of the second polar body (Park et al., 2016). The treated eggs were reared in a 100 L glass aquarium. After two months, a flow-cytometric analysis was performed to estimate the average cellular DNA content of all the triploid individuals. After the fish were anaesthetized with 100 ppm clove oil (Sigma, USA), sample tissues were removed from the tail fins of all individuals for analysis. One million tail fin cells were collected and stained with a high-resolution DNA staining kit (Partec GmbH, Germany) in the dark at room temperature for 15 min. The stained samples were analysed on a Partec PA-II flow cytometer to determine their relative DNA contents. Red blood cells (2.8 pg DNA/nucleus) of the mud loach, Musgurnus mizolepis, were used as the standard reference. Individuals confirmed to be triploid were quarantined in a 30 L glass aquarium for the experiment.
On 16 May 2016, 230 days after hatching, 120 marine medaka were selected for the study, 60 diploids and 60 triploids, with 30 males and 30 females in each group. The diploid and triploid samples were fixed in 10% neutral formalin. To avoid the guts of the sampled fish being distended by large quantities of food, the fish were starved for 24 h before sampling (Park et al., 2001b). On 23 May 2016, fixed samples from each group (n=60) were photographed with a digital camera (Nikon D80, Nikon, Japan). A scale bar was included in the pictures, and they were printed with a laser printer (HP LaserJet 1010, Epson, Japan). Standard length measurements were made from the pictures to the nearest 0.01 cm, using digital Vernier calipers (CD-20CP; Mitytoyo, Japan). Using the methods of Im et al. (2016), body outline measurements were made for 19 distances between landmarks for both truss and classical dimensions (Fig. 1, Table 1). Ls, HHAD, and HHAA indicate horizontal distances, and the other distances (DHAD, DHDC, DHAA, DHAV, DPAA, DADAA, etc.) indicate direct distances (Fig. 1, Table 1).
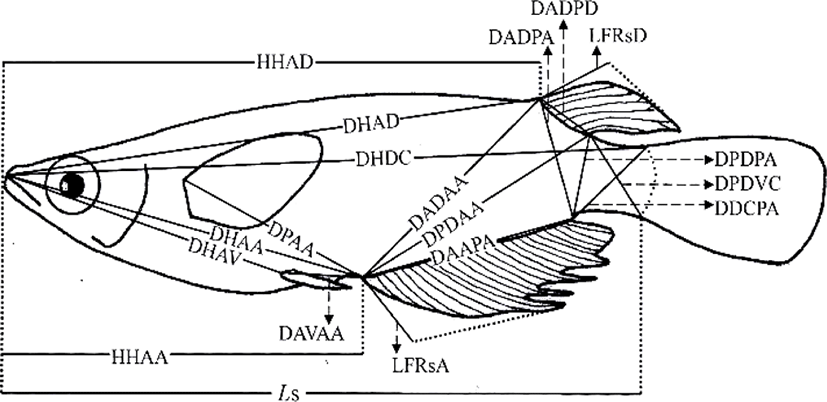
One-way analysis of variance (ANOVA) was used to determine the significance of the differences in various parameters between the diploid and triploid fish (p<0.05, n=60). The most significant variables were then used for a stepwise discriminant analysis (n=60) to determine the maximum separation between the groups and sexes (Bonar et al., 1988). Differences between means were regarded as significant at p<0.01 or p<0.05.
RESULTS AND DISCUSSION
The differences in all 18 morphometric characteristics of the diploid and triploid marine medaka, Oryzias dancena, were analysed with ANOVA, and the results are given in Table 2. This analysis identified differences between the diploid and triploid fish in every characteristic examined, except DADPA, and the most significant differences were observed in DHDC, LFRsD, LFRsA, and DAVAA (p< 0.01). The differences between the diploid and triploid marine medaka in the 18 morphometric characteristics were also analysed by ANOVA separately for the two sexes, and the results are given in Table 3. In this analysis, every morphometric characteristic differed between the diploid and triploid fish when the sexes were assessed separately. The most significant differences between the diploid and triploid males were observed in DHAD, DHDC, DADPD, LFRsD, LFRsA, and DAVAA (p<0.01). The most significant differences in the diploid and triploid females were observed in DHAD, DHDC, LFRsD, LFRsA, and DAVAA (p<0.01).
Refer to the Table 1 for details of the dimensions. There were 60 fish in both the diploid and the triploid samples. ANOVA: **p<0.01, *p<0.05, NS: not significant.
Refer to the Table 1 for details of the dimensions. There were 30 males and 30 females in each ploidy group. ANOVA: **p< 0.01, *p<0.05, NS: not significant.
No classical dimensions were among the characteristics that differed most significantly between the diploid and triploid fish. Classical dimensions have been used to study the characteristics of fish for over 30 years, and these classical dimensions focus on the length, width, and height of the fish, together with the axis of the fish body, including the tail and head (Straüss and Bond, 1990; Park et al., 2004). In this study, we looked for differences in the classical dimensions between the sexes of the marine medaka, but no clear differences were detected. The most significant differences between the two sexes of both ploidies in the truss dimensions were in DHAD, DHDC, LFRsD, LFRsA, and DAVAA. The truss dimensions are a way of dividing the fish body into functional units to investigate different parts of their appearance (Straüss and Bond, 1990). They complement the vertically measured classical characteristics because they measure distances that cross the body shape diagonally, and are also used to determine network type of the body (Park et al., 2004). Both the diploid and triploid male marine medaka had a larger standard length and appearance in the middle of tail that is connected to the body of fish than the females of the same ploidy. The truss dimensions have been used to compare the characteristics of Rhynchocypris oxycephalus under conditions of hunger and satiety (Park et al., 2004). In that experiment, there were changes in the main body region and the region between the rear body and the tail, which reflected the different food available in the different habitats of R. oxycephalus. However, because no difference was observed under the head in this comparison of R. oxycephalus in conditions of hunger and satiety, this measure could be used to identify Rhynchocypris (Park et al., 2001b). In an experiment examining the sexual dimorphism of the Korean chub, the female had a higher index for the direct distance between the insertion of the dorsal fin and the insertion of the anal fin (IDF-IAF) than the male and the height of the female was greater than that of the male (Kim et al., 2008).
Significant differences were observed between diploid and triploid male marine medaka in DHAD, DHDC, DADPD, LFRsD, LFRsA, and DAVAA, and significant differences in the female diploid and triploid fish were observed in DHAD, DHDC, LFRsD, LFRsA, and DAVAA (Table 4). To separate the two groups, the most useful combination of these variables for both males and females was LFRsD, LFRsA, and DAVAA, which correctly classified 85% of each sex as triploid or diploid, with the maximum degree of separation. Table 5 shows the classification function coefficients of the most significant variables in each experimental group. The classification functions (C) in Table 1, developed with stepwise discriminant analysis (Bonar et al., 1988) for male marine medaka were C= 7.269(LFRsA)-5.248(LFRsD)+4.375(DAVAA)-25.151 for diploid marine medaka and C=9.849(LFRsA)-7.262 (LFRsD)+5.112(DAVAA)-28.886 for triploid marine medaka. Assignment of the stock data to one of these two equations resulted in correct classification 85% of the time (Table 6). The classification functions developed with stepwise discriminant analysis for female marine medaka were C=7.157(LFRsA)-5.197(LFRsD)+4.257(DAVAA)-24.257 for diploid fish and C=9.304(LFRsA)-7.349 (LFRsD)+ 5.349(DAVAA)-27.861 for triploid fish. Assignment of the stock data to one of these two equations resulted in correct classification 85% of the time (Table 6).
| Standardized canonical discriminant function coefficients | Function1 | |
|---|---|---|
| Male | Female | |
| DHAD | 0.388 | 0.291 |
| DHDC | 0.412 | 0.387 |
| DADPD | 0.236 | - |
| LFRsD | 1.054 | 0.943 |
| LFRsA | 1.694 | 1.317 |
| DAVAA | 0.949 | 0.756 |
Refer to the Table 1 for details of the dimensions.
Refer to the Table 1 for details of the dimensions.
| Sex | Ploid | Predicted group membership | Total | |
|---|---|---|---|---|
| 2n | 3n | |||
| Male | 2n | 43 (85.7%) | 7 (14.3%) | 50 (100) |
| 3n | 6 (12.7%) | 44 (87.3%) | 50 (100) | |
| Female | 2n | 42 (85.1%) | 8 (14.9%) | 50 (100) |
| 3n | 7 (14.3%) | 43 (85.7%) | 50 (100) | |
Refer to the text for the details of the classification.
Bonar et al. (1988) evaluated the separation of triploid and diploid grass carp by their external morphology, using classification functions based on body depth, gape width, and cheek height. The separation rate was correct only 65% of the time. In our study, the correct separation rate was 85%. To increase the accuracy of this separation, the terms with the lowest discriminant coefficients (DHAD, DHDC, and DADPD) among the most significant variables were excluded, but this did not improve the accuracy of the separation.
The differences in all 18 morphometric characteristics of the male and female triploid fish were analysed by ANOVA, and the results are given in Table 7. The males and females differed in every characteristic, and the most significant differences were observed in DADAA, DPDAA DADPA, LFRsD, and LFRsA (p<0.01). Among the three characteristics DADAA, DPDAA, and DADPA, DADAA differed most significantly between the male and female triploid fish. The lengths of the dorsal fin and anal fin of diploid and triploid male were greater than those of diploid and triploid female, which was also evident to the naked eye (Fig. 2). Ls, DHDC, LFRsD and LFRsA of triploid male and female were longer than those of diploid male and female, but body depth of triploid male and female were shorter than those of diploid male and female, which was evident to the naked eye (Fig. 2).
Refer to the Table 1 for details of the dimensions. There were 30 males and 30 females in the samples. ANOVA: **p<0.01, *p<0.05, NS: not significant.
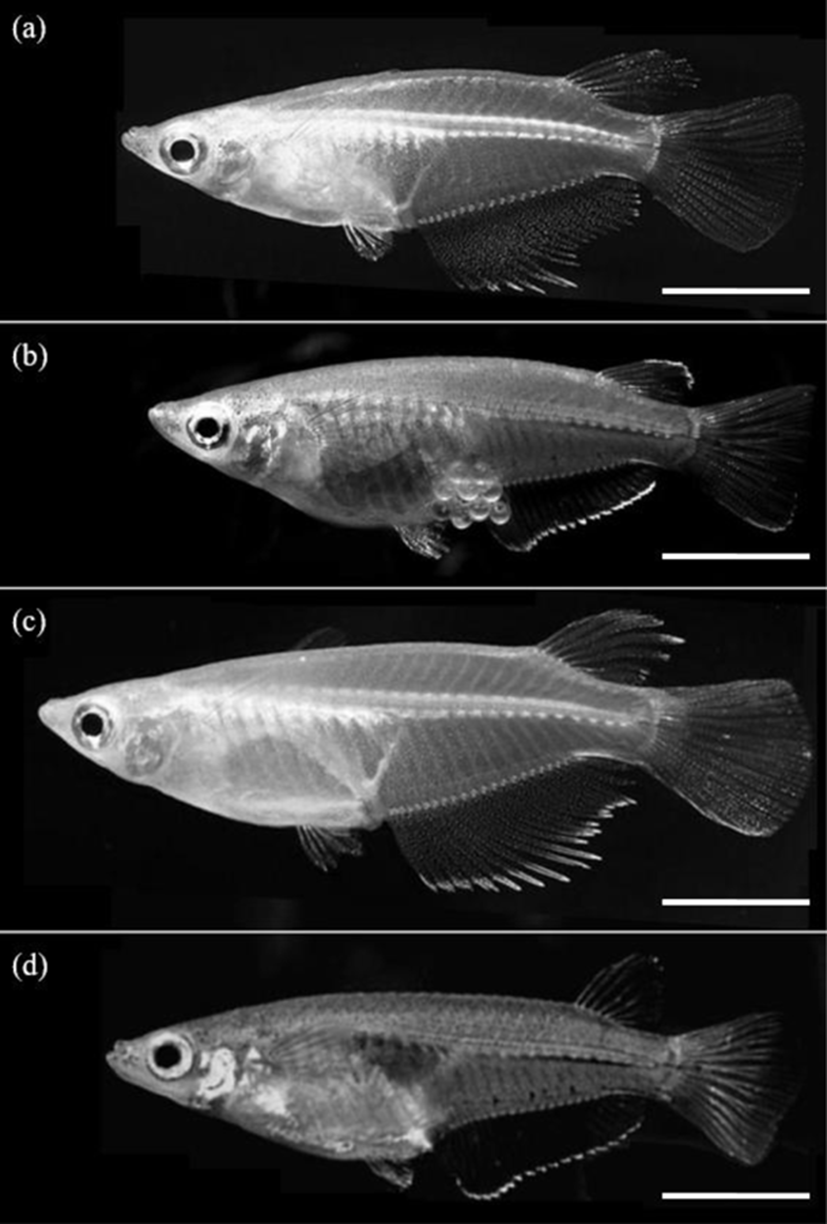
As mentioned by Kim et al. (2009b), it is easy to classify the sexual dimorphism of the marine medaka through the forms of the dorsal and anal fins. Im et al. (2016) also reported that the sexual dimorphism of the diploid marine medaka is evident in the difference in growth 70 days after hatching, with apparent differences in DADAA, DPDAA, DADPA, LFRsD, and LFRsA, with the male characteristics larger than those of the female. The characteristics of the dorsal and anal fins of the marine medaka are significantly different between the males and females. That is, the male marine medaka grows more rapidly than the female, reaching a greater length, with longer dorsal and anal fins. As mentioned by Lim et al. (2012), during spawning, the male marine medaka stacks his anal fin, the anal fin of the female marine medaka, his dorsal fin and then her dorsal fin. Thus, the body of the female is covered by the dorsal and anal fins of the male. Spawning then begins after covering is completed (Lim et al., 2012). For this reason, the lengths of the dorsal and anal fins of the male are greater than those of the female. Measurement of the characteristics of the two sexes of the cocktail wrasse, Pteragogus aurigarius, by Park et al. (2001a) showed significant differences in the lengths of the first and second fin rays of the dorsal fin according to sex.
External morphological variations are a component of sexual dimorphism, together with features such as the genital papilla, body pigmentation, and fin shape (Kim & Kim, 2001). Sexual dimorphism is the most conspicuous difference between the sexes (Kim et al., 2008), and occurs in many fishes. Females are usually larger than males of the same age, although in some species, males are larger than females, e.g., the gudgeon, Gobio gobio (Mann, 1980), and the filefish, Brachaluteres ulvarum (Akagawa et al., 1995). The reasons for these size differences are unclear (Katano, 1998). Several authors have reported that the evolution of a larger body size in males probably results from male-male competition associated with a polygynous mating system (Katano, 1998; Kim et al., 2008). Therefore, exploring the nature and extent of sexual dimorphism can extend our understanding of social structure and adaptation, as well as species identification.
There are a number of techniques for separating diploid and induced triploid fishes (Thorgaard et al., 1982; Wolters et al., 1982; Park et al., 2016). The three most widely accepted methods now used are flow cytometric measurements of erythrocyte DNA (Thorgaard et al., 1982; Park et al., 2016), Coulter counter estimation of erythrocyte nuclear size (Thorgaard et al., 1982; Park et al., 2016), and measurement of erythrocyte nuclear volume by light microscopy (Wolters et al., 1982; Park et al., 2016). Unfortunately, the first two methods are very expensive, and light microscopy is time-consuming and insufficiently accurate for widespread use. Clearly, any method that would be as accurate as, more rapid, and less costly than these three techniques would be helpful to marine medaka researcher.
In this study, when their classical dimension and truss dimension was measured, triploid marine medaka were shown to grow more rapidly than diploid marine medaka, with sexual dimorphism expressed as longer dorsal and anal fins in the males. The differences in these characteristics should be useful in experiments in which the ploidy of the marine medaka must be established. These results extend our knowledge of the morphometric changes that occur in diploid and triploid marine medaka and may be useful for application to the sterility of the marine medaka in the living modified organism evaluation project.
