INTRODUCTION
Follicle stimulating hormone (FSH) belongs to the glycoprotein hormone family, which includes luteinizing hormone (LH), chorionic gonadotropin (CG), and thyroid stimulating hormone (TSH) (Min et al., 2004). FSH is a central hormone of mammalian reproduction, necessary for gonadal development and maturation at puberty and for gamete production in the ovary and testis during the fertile phase of life (Simoni & Nieschlag, 1995). Thus, the role of FSH in regulating the gonadal function has been well-documented. Intact ligand-receptor interaction is needed for normal hormone action, and an anomaly in the structure of either the gonadotropin ligand or its receptor may cause disturbances in reproductive functions (Tapanainen et al., 1998). FSH acts by binding to specific receptors localized exclusively in the gonads (Simoni et al., 1997).
G protein-coupled receptors (GPCRs) relay information from extracellular stimuli to intracellular responses in a wide range of physiological and pathological processes (Martemyanov & Carcia-Marcos, 2018). Recently, mini G proteins have proven to be versatile tools for studying GPCR activation and coupling specificity in cells, and are useful for discovering and characterizing G protein subtype-biased ligands (Wan et al., 2018). The gonadotropin receptors belong to a large superfamily of GPCRs characterized functionally by their interaction with guanine nucleotide-binding proteins and structurally by their seven transmembrane spanning domains, extracellular amino-terminus, and intracellular carboxy-terminus. Like other GPCRs, agonist-induced FSH receptor (FSHR) activation and phosphorylation are now recognized as important steps in agonist-induced internalization of the FSHR (Nakamura et al., 2000). Most of the internalized human CG/rat LH receptor (hCG/rLHR) complexes is routed to the lysosome where the hormone and the receptor are degraded (Kishi et al., 2001). However, hFSH/hFSHR complex accumulates in endosomes and is subsequently recycled back to the cell surface where the bound, intact hFSH dissociates back into the medium and can bind to the receptor again (Krishnamurthy et al., 2003). The recycling or degradation of the internalized gonadotropin receptors results in the maintenance or loss of cell surface receptors, respectively (Kishi et al., 2001; Galet et al., 2003, 2004; Hirakawa & Ascoli, 2003). The receptor recycling in hLH/CGR and hFSHR promotes the maintenance of cell surface receptors and preserves hormonal responsiveness (Bhaskaran & Ascoli, 2005).
Analysis of the truncation of the rLH/CGR, the phorbol ester phorbol 12-myristate-13-acetate (PMA), or agonist-induced phosphorylation of this receptor maps to four serine residues in the C-terminal cytoplasmic tail. The agonist-induced phosphorylation is completely abolished in rLH/CGR truncated at residue 631 (Hipkin et al., 1995a). The residues 632-53 in the C-terminal tail of the rLH/CGR are involved in PMA- and hCG-induced desensitization and down-regulation (Wang et al., 1996). rFSHR was truncated at residue 635, removing all but one of the potential phosphorylation sites present in the C-terminal tail. The truncated FSHR bound to hFSH with the appropriate affinity and responded with increases in cAMP and inositol phosphate accumulation (Hipkin et al., 1995a). Thus, rLH/ CGR prevented phosphorylation by the truncation of the C-terminal cytoplasmic tail and prevented gonadotropin- or PMA-induced uncoupling (Ascoli, 1996). The post-endocytotic fate of the gonadotropin receptors is an important determinant of the desensitization of gonadotropin responses (Bhaskaran & Ascoli, 2005).
To better define the involvement in signal transduction of recombinant eel follicle-stimulating hormone (rec-eelFSHβ/α)-stimulated cAMP synthesis, we constructed a mammalian expression vector of eel follicle-stimulating hormone receptor (eelFSHR) and truncated the eelFSHR C-terminal phosphorylation sites. We investigated signal transduction in cells expressing truncated and normal receptors in CHO-K1 cells and PathHunter Parental cell lines expressing β-arrestin.
MATERIALS AND METHODS
The mammalian expression vector, pcDNA3, CHO-K1 suspension (CHO-S) cells, FreeStyle MAX reagent, FreeStyle CHO expression medium, pCMV-ARMS1-PK2 expression vector, antibiotics, and AssayComplete medium were purchased from Invitrogen Corporation (San Diego, CA, USA). The following reagents and materials were also used: restriction enzymes and a DNA ligation kit (Takara, Tokyo, Japan); CHO cells (Japanese Cancer Research Resources Bank, Tokyo, Japan); and Ham's F-12 medium, Opti-MEM I, serum-free CHO-S-SFM II, Geneticine, and Lipofectamine 2000 (Gibco BRL, Grand Island, NY, USA). Fetal bovine serum (FBS) was obtained from Hyclone Laboratories (Logan, UT, USA). Pro-PrepTM protein-extraction solution was obtained from Intron Biotechnology (Seoul, Korea). Lumi-Light western blot kit was purchased from Roche (Basel, Switzerland). PathHunter CHO-K1 β-arrestin Parental cell line was obtained from DiscoveRx (San Diego, CA, USA). Disposable spinner flasks and grass flasks were obtained from Corning Incorporated (NY, USA). The cAMP Dynamic 2 immunoassay kit was purchased from Cisbio Bioassay (Codolet, France). All other reagents used were obtained from Sigma-Aldrich Corp (St. Louis, MO, USA). The oligonucleotides used in this study were synthesized by Genotech (Daejon, Korea).
To obtain the mammalian expressing vector, the cDNA encoding the full-length eelFSHβ-subunit was fused with the mature protein portion of the α-subunit using overlapping polymerase chain reaction (PCR) mutagenesis, as previously reported (Kim et al., 2016a,b). These PCR fragments were digested with EcoRI and SalI enzymes and then ligated into the EcoRI and XhoI sites of the eukaryotic expression vector pcDNA3 (designated as pcDNA3-eelFSHβ/α). The schematic diagram of rec-eelFSHβ/α is shown in Fig. 1. The orientation of the insert was confirmed through restriction mapping. Finally, this vector was sequenced completely to confirm the Kozak site and absence of PCR-induced errors.
The rec-eelFSHβ/α proteins were expressed by transfecting the expression vector into CHO-S cells using the FreeSytle MAX reagent transfection method, according to the supplier's instructions and as described previously (Lee et al., 2017; Byambaragchaa et al., 2018). Briefly, CHO-S cells were cultured with FreeStyle CHO expression medium at 1×107 cells/ 50 mL for 3 days. One day prior to transfection, the cells were passaged at 5-6×105 cells/mL with 50 mL CHO expression medium in disposable spinner flasks. On the day of transfection, the cell density should be approximately 1.2-1.5×106 cells/mL. Next, DNA (40 μg) was mixed gently in 1.2 mL of OptiPRO serum free medium (SFM), and FreeStyle MAX reagent (40 μL) for transfection was mixed gently in 1.2 mL of the OptiPRO serum free medium. Both mixed media were incubated for 5 minutes at room temperature (RT). Then, the complex (2.4 mL) was added to each cell suspension flask. For the rec-protein assay, 2 mL of culture medium was collected on days 1, 3, 5, and 7. Finally, the culture media were collected on day 7 after transfection and centrifuged at 15,000 rpm at 4℃ for 10 min to remove cell debris. Supernatants were collected and frozen at -80℃. The samples were then concentrated by freeze-drying and mixed with PBS. The rec-protein was analyzed by western blot and enzyme-linked immunosorbent assay (ELISA).
The rec-hormones were quantified by ELISA using anti-eelFSH α- and β-subunit monoclonal antibodies and enzyme conjugate coupled to horseradish peroxidase and TMB substrate as previously developed in our lab (Kim et al., 2016a). The samples in culture media (100 μL) collected on days 1, 3, 5, and 7 were analyzed. After the freeze-dry process, the concentrated sample was also mixed with PBS. The 1st antibody (eFA5) was coated in 96-well plates and then incubated overnight at 4℃. After blocking with 0.5% casein in PBS for 1 h at 37℃, dilution samples of antigen (0-800 ng/mL) and an undiluted solution of rec-eelFSH β/α were added and incubated for 1 h at 37℃. After washing 3 times with PBS-T, HRP-conjugated anti-eel monoclonal antibody (eFA11) diluted 100-fold in PBS was added and incubated for 1 h at RT, and after washing, 50 μL of tetramethylbenzidine substrate was added to each well and the plates were incubated at RT in the dark. After 15-30 min, the reaction was terminated by adding 50 μL of a stop solution (20% H2SO4) to each well, the plate reader determined the absorbance at 450 nm.
For western blot analysis, the concentrated sample (10 μg) was subjected to reducing 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli (1970). After SDS-PAGE, the proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (0.2 μM) using a Bio-Rad Mini Trans-Blot electrophoresis cell as previously reported (Lee et al., 2017). The membranes were washed with 1x Tris-Buffered Saline and Tween 20 (TBS-T) and incubated with primary antibody (anti-eelFSHβ/α) diluted 1:1,500. Next, the membranes were reacted with the secondary antibody (goat anti-mouse IgG-HRP) diluted with 1:3,000. Subsequently, membranes were incubated for 1 min with 2 mL of the Lumi-Light substrate solution and exposed on X-ray film for 1-10 min.
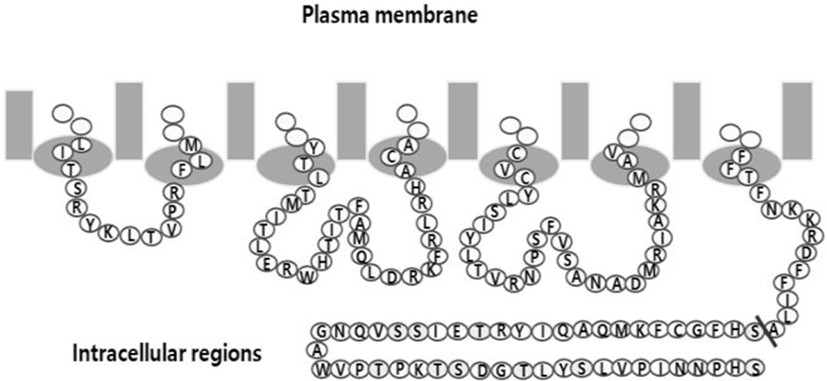
eelFSHR cDNA was cloned using cDNA of eel testis and ovary, as previously reported (Byambaragchaa et al., 2018). The PCR fragments were ligated into the pcDNA3 mammalian expressing vector by the XhoI and EcoRI enzyme sites (designated as pcDNA3-eelFSHR-WT). For the Parental cells, the NheI- and SacI-digested eelFSHR cDNA was cloned into the same enzyme sites as the pCMV-ARMS1-PK2 expression vector (designated pCMV-ARMS1- PK2-eelFSHR-WT). As shown in Fig. 2, cDNAs encoding for eelFSHR truncated at residue 614 (designated pcDNA3- eelFSHR-t614; pCMV-ARMS1-PK2-eelFSHR-t614) were constructed using the polymerase chain reaction (PCR) by introducing a stop codon after the codon for amino acid residue alanine. There is no stop codon in the C-terminal region of SacI enzyme site. The orientation of the insert was confirmed through restriction mapping. The identity of this construct was verified by sequencing the entire open reading frame.
Transfections of CHO cells were performed using the liposome transfection method as previously described (Park et al., 2010). The transfected cells were adjusted for cAMP analysis at 48-72 h after transfection. PathHunter CHO-K1 EA-Parental cells, which are engineered to stably express the enzyme acceptor-tagged β-arrestin fusion protein, were transiently and stably transfected according to the supplier's protocol. For the stable cell lines, the transfected cells were seeded with 500 and 1,000 cells in 100 mm culture dishes at 24-48 hr after transfection. The cells were cultured in AssayComplete medium containing G418 for 2-3 weeks to isolate the cells expressing eelFSHR. Approximately 20 clones were recovered and cultured in a 24-well plate. The grown cells were transferred to 6 well plates and 25 cm2 culture flasks. Finally, 5 cell clone lines were isolated and stocked according to the method previously reported (Lee et al., 2017).
Measurement of AMP accumulation in CHO cells and PathHunter CHO-K1 EA-Parental cells was performed using cAMP Dynamics 2 competitive immunoassay kits (Cisbio Bioassays) as described previously (Byambaragchaa et al., 2018). The standard samples were prepared to cover an average range of 0.17-712 nM (final concentration of cAMP per well). After the cells (10,000) were seeded into 384-well plates and 5 μL compounds in medium and buffer were added to each well. The plate was sealed and incubated for cell stimulating at RT for 30 min. Then, cAMP-d2 (5 μL) and anti cAMP-cryptate (5 μL) were added to each well. After, the plate was sealed and incubated at RT for 1 hr. The absorbance of each well was read on a compatible HTRF reader. Results are calculated from the 665 nm /620 nm ratio and expressed as Delta F % (cAMP inhibition).
The cAMP concentration for Delta F% values was calculated by GraphPad Prism 6.0 (GraphPad Software, Inc, La Jolla, CA, USA).
Dose-response curves were fitted with a nonlinear regression, variable slope equation using GraFit 5.0 (Erithacus Software Limited, Surrey, UK) and GraphPad Prism 6.0. Curves fitted in a single experiment were normalized to the background signal measured for mock-transfected cells (0%). Each sum curve was calculated from at least three independent experiments.
RESULTS
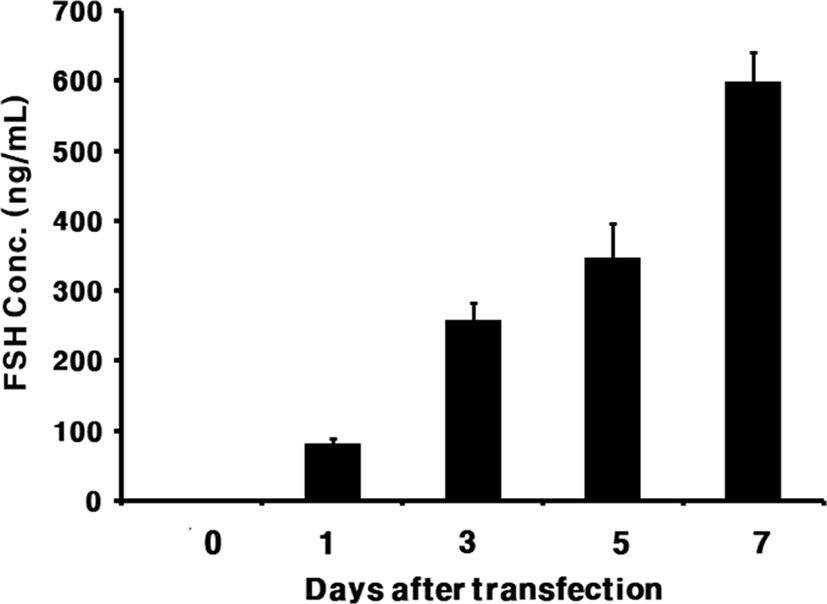
The level of rec-eelFSHβ/α produced is shown in Fig. 3. The secreted quantity in the media was approximately 88.1±4 ng/mL on day 1 after transfection. The expression quantity gradually increased and was detected approximately 347.4±13 ng/mL on day 5 after transfection. The expression level was high, at 598.5±30 ng/mL, on day 7.
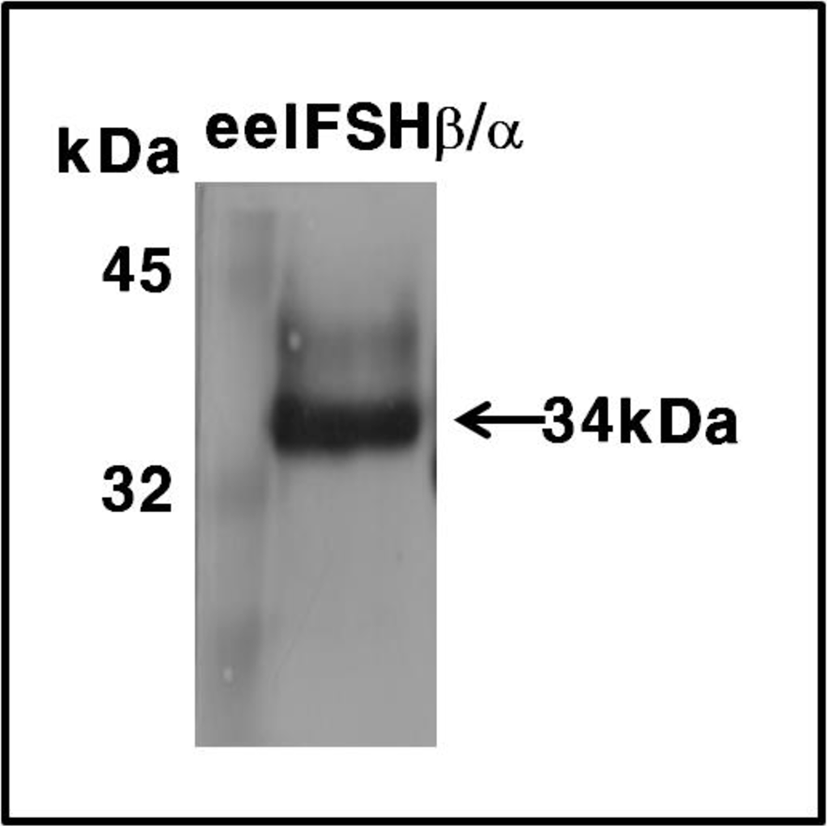
These expression levels were 2 times higher than that of the transient expression in attached CHO-K1 cells. The molecular weight of the rec-eelFSHβ/α protein was approximately 34 kDa. The band was detected broadly as shown in Fig. 4. Thus, we suggest that oligosaccharides of about 10 kDa were added to the rec-eelFSHβ/α produced in the CHO-S cell system.
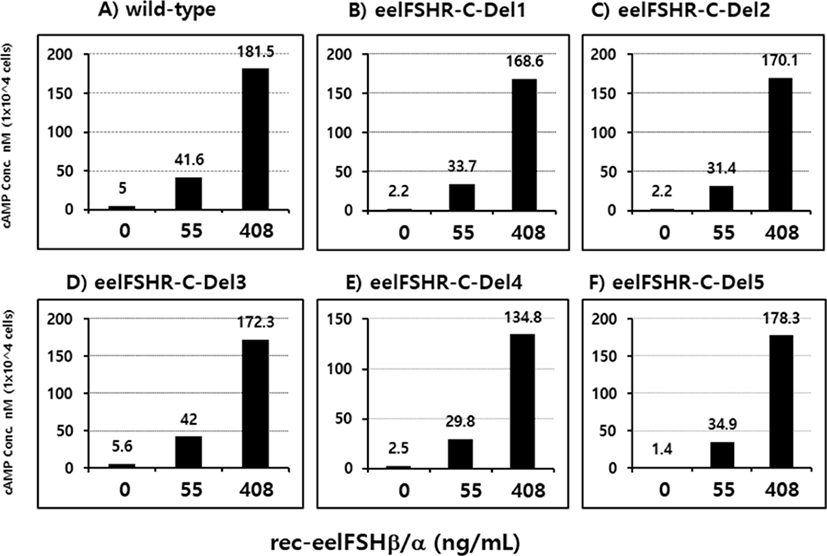
PathHunter-EA CHO Parental cells transfected with eelFSHR cDNA were isolated by G418 treatment. Stable clones were selected by G418 treatment. 5 clones were subjected to cAMP analysis by rec-eelFSHβ/α (0, 55, and 408 ng/mL). As shown in Fig. 5, cAMP responsiveness was the highest in two clones (Del 3 and Del 5).
The effect of rec-eelFSHβ/α on cAMP stimulation was determined to evaluate the activity in CHO-K1 cell lines expressing both the eelFSHR-WT and eelFSHR-t624 genes. Receptor cells were incubated with dose-dependent concentrations (0.008-405 ng/mL) of rec-eelFSHβ/α. Here, cAMP production expressed as Delta F%, was inhibited by the activation of the transfected eelFSHRs (data not shown). Then, these data were calculated by cAMP concentration (nM), as shown Fig. 6A. The cAMP concentration increased in direct proportion to the concentration of the rec-eelFSHβ/α. The EC50 values in both eelFSHR-WT and eelFSHR-t624 were 37.3±2 ng and 61.8±5 ng, respectively (Table 1). The EC50 value following C-terminal deletion in CHO-K1 cells was about 60.4% of that of eelFSHR-WT. The maximal responses of eelFSH-WT and eelFSHR-t614 were 290.4±14 and 189.9±15 nM, respectively. The maximal response was greatly decreased by C-terminal deletion of eelFSHR. Thus, the truncation of the C-terminal cytoplasmic tail phosphorylation sites plays a pivotal role in cAMP responsiveness (EC50 value) and maximal response in CHO-K1 cells.
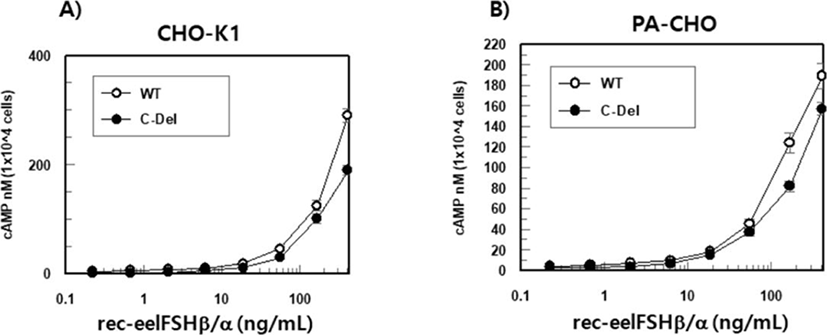
As shown in Fig. 6B, the EC50 values calculated by cAMP stimulation in both cells were 53.0±4 and 46.8±6 ng, respectively (Table 1). The maximal responses of eelFSH-WT and eelFSHR-t614 were 203.9±19 and 157.8± 8 nM, respectively. As shown in CHO-K1 cells (Fig. 6A), the maximal response with eelFSHR-t614 was lower than eelFSHR-WT.
Values are the means±SEM of triplicate experiments. The EC50 values used to determine the potencies were determined from the concentration-response curves for the in vitro bioassays.
rec-eelFSHβ/α, recombinant eel follicle-stimulating hormone; eelFSHR-WT, eel follicle-stimulating hormone receptor wild type.
DISCUSSION
GPCRs signal by coupling to heterotimeric G proteins and arrestins, which alternately activate kinases (βARKs, GRKs) and other appropriate molecules to control virtually every cell function. These second messenger-independent kinases further phosphorylate the agonist-induced receptors on serine and/or threonine residues located in the C-terminal region of GPCR. The experiments presented herein were designed as an initial attempt to identify the roles of the phosphorylated residues and their responsibility for the uncoupling of eelFSHR from adenylyl cyclase.
Our results showed that rec-eelFSHβ/α was efficiently secreted into the medium in the CHO-S cells. The expression level was highly detected on day 7 after transfection. The size of the western blot was approximately 34 kDa. We also established stable cell lines of eelFSHR-C-Del in the PathHunter-EA Parental cells expressing β-arrestin, which function as adaptor proteins specifically targeting GPCRs for dynamin-dependent endocytosis via clathrin-coated vesicles (Mokrosinski et al., 2012). These results suggest that rec-eelFSHβ/α show activity in cells expressing eelFSHR.
In the analysis of the secretion of rec-glycoprotein hormones, we have also previously reported on the activities of equine CG, equine FSH, and hCG in attached CHO cells (Saneyoshi et al., 2001; Min et al., 2004; Park et al., 2009; Jeoung et al., 2010; Park et al., 2010, 2017;). Our results are consistent with those of earlier studies that demonstrated rec-eCGβ/α in CHO-S cells could be translated and efficiently secreted as its biologically active tethered form (Min et al., 2004; Lee et al., 2017). rec-eCGβ/α showed almost the same pattern of expression from 1 to 7 days after transfection. However, rec-eelFSHβ/α expression in our study was very low on day 1 after transfection, but gradually increased, showing the highest expression on day 7 after transfection. We suggest that these results indicate the purpose of the O-linked oligosaccharide chains in the C-terminal region of the eCGβ-subunit, as without the C-terminal region, rec-eelFSHβ/α was secreted at a low level on day 1 after transfection.
In the western blot results, the band was broadly detected at 34 kDa. These results are consistent with previous studies of single-chain rec-eCGβ/α which suggest an approximate size of 43-45 kDa in attached CHO cells (Park et al., 2009, 2010, 2017). rec-eCGβ/α expressed in Sf9 insect cells appears at ~45 kDa and heterodimeric eCG is present with an upper band at 45 kDa and a lower ban at ~38-40 kDa (Legardinier et al., 2008). Thus, glycoproteins including eelFSHβ/α and eCGβ/α in mammalian cultured cells (CHO-K1, COS7, and CHO-K1 suspension cells) were consistent in modified with the molecular weight.
In the presented study, our results indicate that the EC50 value of a mutant FSHR cDNA with a truncated C-terminal cytoplasmic tail was about 60.4% of the eelFSHR-WT EC50 in CHO-K1 cells. The maximal response was greatly decreased by C-terminal deletion in eelFSHR. The cAMP response of the truncated eelFSHR in PathHunter Parental cells was also somewhat decreased compared to eFSHR-WT, as shown in the graph, but the EC50 value was slightly increased.
In the rat FSHR (rFSHR), the response of a deletion mutant of the C-terminal cytoplasmic tail in the residue 635 (FSHR-t635) is higher than those of cells expressing the FSHR-WT, but the concentration of hFSH required to elicit these responses is similar in both cell lines (Hipkin et al., 1995a). The study suggested that the potency of hFSH is similar in cells expressing FSHR-WT or FSHR-635, but the efficacy of hFSH is 2-3-fold higher in cells expressing FSHR-t635. It also suggests that hFSH can both phosphorrylate and uncouple in the FSHR-t635. However, other groups have reported that the C-terminal cytoplasmic tail of the rLH/CGR (rLH-t631) is necessary for agonist-induced uncoupling (Sanchez-Yague et al., 1992). The truncation of the C-terminal cytoplasmic tail of the β2-adrenergic receptor causes a delay in the onset of agonist-induced uncoupling (Bouvier et al., 1988).
In rLH/CGR, hCG and phorbol 12-myristate-13-acetate (PMA) increase receptor phosphorylation in the wild type receptor and the receptor truncated at residue 653, but not in the receptor truncated at residue 631 (Hipkin et al., 1995b). The C-terminal tails of rLH/CGR truncated at residues 632-653 is involved in PMA-induced desensitization, hCG-induced desensitization, and hCG-induced down- regulation (Wang et al., 1996). These results are similar to those reported for several C-terminal truncations of the LH/CGR (Rodriguez et al., 1992). Thus, some truncations of the C-terminal cytoplasmic tail in rLH/CGR prevent phosphorylation and retard or prevent gonadotropin- or PMA-induced uncoupling. However, an equivalent truncation of the rFSHR does not affect phosphorylation or uncoupling (Ascoli, 1996). In the present study, our data suggest that the truncation of the C-terminal cytoplasmic tail of eelFSHR (eelFSHR-t614) greatly decreased the eelFSH-induced cAMP responses in cells. Thus, we propose that these data are not consistent with rFSHR being truncated in the C-terminal cytoplasmic tail, but are consistent with the truncation of rLH/CGR and β2-adrenergic receptor. Thus, we suggest that the C-terminal phosphorylation sites in the eelFSHR are necessary in cell types expressing β-arrestin. Further studies are required to elucidate the functional mechanisms that regulate the roles of hormone receptors in eel ovaries and testes during sex-maturation.
