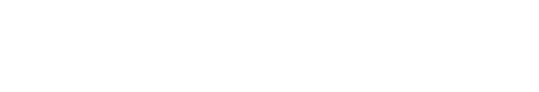INTRODUCTION
Loach (Misgurnus anguillicaudatus) is a demersal freshwater teleost, widely distributed in southeastern Asia, including China, Japan and Korea. The loach can be used as a traditional medicine or folk remedy for treatments of hepatitis, osteomyeitis, carbuncles, inflammations and cancers, as well as for patient’s recovery from debilities caused by various pathogens and aging (Qin et al., 2002). It is also recognized as economically important species, with a gradually increasing market demand in recent years. However, loach aquaculture is negatively affected by early life stage mortality, which attracted our interest to characterize how their immune system works during the development stages. In fishes, larvae are immediately exposed to various pathogens after hatching (Zapata et al., 1997). However, immune system of young fish is still developing and not all of the structures and functions present in the adults are in the larvae (Ellis, 1988; Tatner, 1996). Therefore, knowledge of the immune system in ontogeny can offer new protective strategies and indicate optimal vaccination point (Vadstein, 1997).
Previous studies show that the expression of immunerelevant transcripts during embryonic and larval development in other fish (Seppola et al, 2009; Ruangsri et al., 2012). However, our understanding of immunity in loach of early life stages is still incomplete, and the information available is restricted to a few immune-related genes. Shen et al (2011) studied the developed 21 new genes and obtained partial coding sequence in M. anguillicaudatus (Shen et al., 2012), and out of these genes, recombinationactivating gene 1 (RAG-1) and sacsin (SACS) have been considered as immune-related genes.
The RAG-1 gene has been reported as very useful markers of the physiological maturity of the immune system (Corripio-Miyar et al., 2007; Zhang et al., 2009; Fan et al., 2009). This gene is essential in the differentiation of immature B and T cells and is expressed in primary lymphoid organs, although not in mature cells (Nagaoka et al., 2000). It is this expression during the maturation of the lymphoid organs that makes the RAG-1 gene for study of the developmental stages in several fishes (Willett et al., 1997; Huttenhuis et al., 2005; Corripio-Miyar et al., 2007; Mao et al., 2012; Covello et al., 2013). The SACS gene that is associated with nervous system diseases in human, whose mutations cause childhood-onset autosomal recessive spastic ataxia of Charlevoix-Saguenay (Kozlov et al., 2011). It is highly expressed in the central nervous system, and is also found in the skin, skeletal muscles, and at low levels in the pancreas. Also, recent study have shown that sacsin protein functions as a molecular chaperone (Anderson et al., 2011). Previously, several studies was observed that virus responsive in fish (Workenhe et al., 2010; Hori et al., 2012), up-regulation of SACS after in vivo stimulation with the viral mimic pIC or challenge with nodavirus (Rise et al., 2010).
The present study aimed to improve knowledge about expression of RAG-1 and SACS in loach, which can provide crucial information regarding the ontogeny of the immune system, with implications for vaccination regimes. Although the RAG-1 and SACS gene has been reported in several fish, considerably less is known about the expression of both genes in loach. Therefore, we investigate the expression pattern analysis in larvae and tissue-specific expression in immune organ.
MATERIALS AND METHODS
Larvae and adult loach was collected from Inland Aquaculture Research Center, National Fisheries Research and Development Institute (NFRDI; Changwon, Republic of Korea), and maintained in 5 tons flow through tank at 24±1°C under a natural photoperiod. The tissue samples was prepared from various tissues including brain, muscle, fin, eye, liver, gill, kidney, and spleen obtained from healthy loach (n=3). Eggs were collected from different developmental stages of embryo including unfertilized oocyte (U), fertilized egg (F), 1-4 cells (E1), 8-64 cells (E2), molura (E3), blastula (E4), gastrula (E5), neurula (E6) and lavae were collected from eight laval stages including 0, 5, 10, 15, 20, 25, 30 and 40 days post hatching (dph) fish kept at 24.0°C in the fresh-water tank. All the samples were collected in Trizol Reagent (Invitrogen) aseptically and preserved at –80°C until RNA extraction.
For the developmental series and unfertilized eggs, each sample was homogenized in Trizol Reagent (Invitrogen) using a motorized Kontes RNase-Free Pellet Pestle Grinder, a disposable nuclease-free plastic pestle and a 1.5 mL microcentrifuge tube, as per manufacturer’s instructions, and suspended in DEPC-treated water for RT-PCR. Total RNA integrity was verified by 1.2% agarose gel electrophoresis, and purity was assessed by A260/280 and A260/230 NanoDrop UV spectrophotometry for both the crude and column purified RNA extracts. cDNA synthesis was used for reverse transcribed into cDNA using First Strand cDNA synthesis kit (Roche). All the primers were designed using bundle software, Primer express (version 3.0). The gene specific primers are listed in Table 1.
To evaluate mRNA levels of maRAG-1 and maSACS, primers were specifically designed to detect and quantify cDNA sequences without detecting genomic DNA. The real-time PCR reactions were monitored with melting curve analysis using 7500 software (version 2.0.5). Amplification efficiency was determined by serial dilutions. All experiments were repeated in tri-replicate. Each reactions displayed an efficiency 92.1% (R2=0.965), 90.2% (R2=0.947) and 98.4% (R2=0.998), maRAG-1, maSACS and β-actin, respecttively.
Real-time PCR reaction mixture contained 2 μL of diluted cDNA sample, specific primer and the Power SYBR Green PCR Master Mix (Applied Biosystems). The reactions for the amplification of each genes was subjected to an initial denaturation at 95°C for 15 sec, annealing at 60°C for 1 min, and elongation at 60°C for 30 sec. Thermal cycling and fluorescence detection were conducted using the 7500 Real-Time PCR system (Applied Biosystems).
Expression of both genes was normalized to an endogenous reference β-actin and presented as subtraction of target CT values from β-actin CT values (ΔCT value). Comparison of gene expression between tissues and calibrator was derived from subtraction of the calibrator ΔCT values from the target ΔCT values to give a ΔΔCT value, and relative gene expression was calculated to determine fold difference (2–ΔΔCT).
The data are presented as the mean ± standard deviation. The mRNA levels of analyzed genes were expressed as a ratio to those β-actin, which was simultaneously amplified as an internal control for each cDNA. The data was statistically analyzed by one-way ANOVA after arcsine transformation when needed, and followed by a Tukey’s test for identification of the statistically distinct groups. Significant differences were accepted for P <0.05.
RESULTS
In previous study, RAG-1 mRNA was expressed in various tissues including the liver, heart, gill, thymus, muscle, brain, intestine, spleen, kidney and gonad, and detected throughout the early larvae period (Mao et al., 2012). The result showed that RAG-1 transcript level was mainly detected in lymphoid organs including the kidney and thymus in red-spotted grouper. Also, recent study have shown that SACS mRNA expressed in nervous system in Atlantic cod (Rise et al., 2010). Such results are now confirmed by the detection of both genes in various tissues by real-time PCR analysis in loach.
In order to investigate of maRAG-1 and maSACS mRNA expression in various tissues from healthy adult loach (2 year-old), oligonucleotide specific primers were designed according to previously reported both genes in loach (GenBank accession no. EF056344 and JN980038, respectively) (Table 1). Gene expression levels were normalized with respect to β-actin (GenBank accession no. AB200265) and spleen mRNA was employed as calibrator. maRAG-1 was expressed ubiquitously in transcript level in all tissues but significantly expressed in intestine and muscle (13.2- and 17.6-fold higher than in the spleen, respectively) (Fig. 1). In contrast, maSACS was highly expressed in brain and eye (23.7- and 16.7-fold higher than in the spleen, respectively) (Fig. 1).
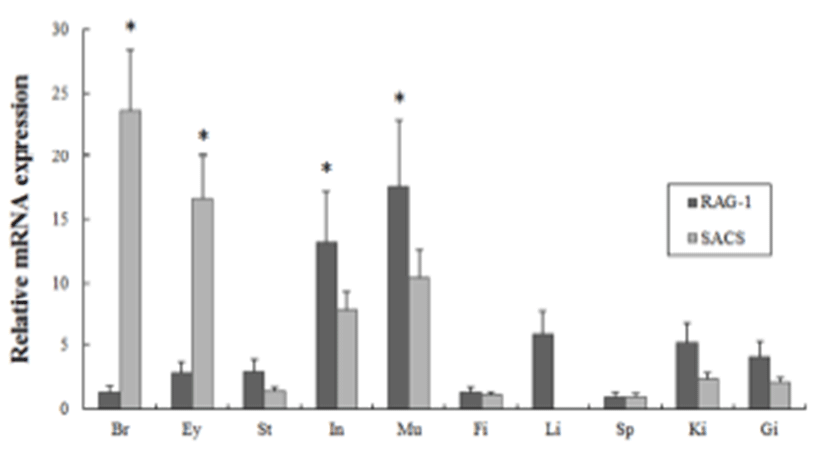
All samples were technically tri-replicated samples, and target genes were standardized using reference gene β- actin for mRNA expression study. The expression pattern of the maRAG-1 and maSACS genes were investigated over time course U, F, E1 to E6 (egg stages) and 0 to 40 days post hatching (larval stages) of loach using real-time PCR.
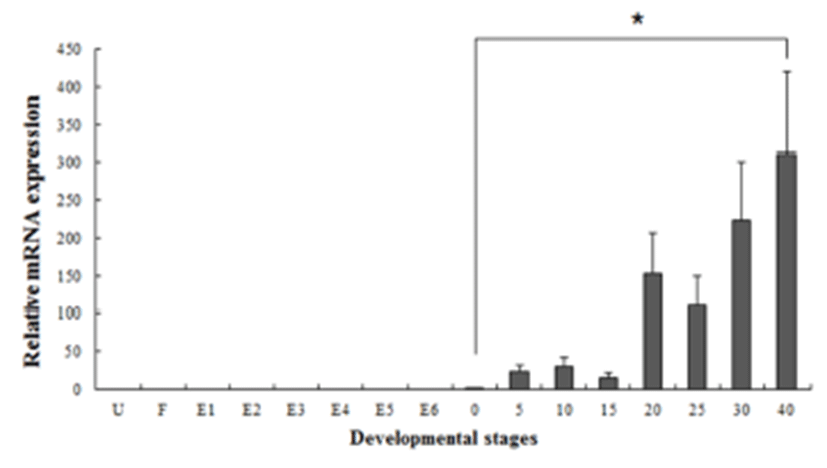
Several studies have reported that some genes are transferred from the mother to the offspring during early period (Zapata et al., 2006). In our study, the maSACS mRNA transcripts were detected in unfertilized oocytes (U), but not detected in maRAG-1 (Fig. 2 and 3). maRAG- 1 mRNA transcripts were first detected at 0 dph (100% hatched), and the transcription level was drastically increased after the 15 dph. The intensity of maRAG-1 transcript was measured on the basis of 0 dph (1.0-fold), each value has the 20 dph (153-fold), 25 dph (112-fold), 30 dph (223- fold), and 40 dph (311-fold), respectively (Fig 2). maSACS expression was highly detected at unfertilized oocyte (U) (50-fold), and significantly decrease until around blastula (E4). After hatching, transcription level was significantly increased in 0 to 40 dph (Fig. 3).
DISCUSSIONS
During embryonic and early larval development, the fish immune system is not fully developed and the immunological capacity is thus assumed to be limited. This study is the first to report the mRNA expression of the important immune genes maRAG-1 and maSACS in loach. In addition their expression was followed from unfertilized oocytes through to the early developmental stage. As both genes are important immune and disease related function, their level of expression can help from the problem of high mortality in larvae. In fish, the RAG-1 mRNA level was first examined in lymphoid organs of rainbow trout larvae (Hansen & Kaattari, 1995), and also investigated in the same organs of zebrafish (Willett et al., 1997) and common carp (Huttenhuis et al., 2005). SACS mRNA level was examined in Atlantic cod larvae, and also profiled of virusresponsive and putative antimicrobial peptide encoding transcript (Rise et al. (2012)).
In our study, maRAG-1 and maSACS was investigated in various tissues in adult loach, including brain, eye, stomach, intestine, muscle, fin, liver, spleen, kidney and gill (Fig. 3). The results are demonstrated that maRAG-1 was ubiquitously expressed in most of the tested tissues, and significantly expressed in mucosal organs including intestine and muscle. maSACS was highly expressed in brain and eye by real-time PCR. Several studies showed that RAG-1 transcript level was mainly detected in lymphoid organs including spleen and kidney (Hansen & Kaattari, 1995; Willett et al., 1997; Huttenhuis et al., 2005; Mao et al., 2012). However, expression of the RAG-1 gene has also been observed in other organs apart from the lymphoid organs, such as pancreas or intestine, with the latter also related to mucosal immunity (Danilova & Steiner, 2002). The observed expressed in the mucosal organs may suggest that rearrangement is occurring in the B cells present in the mucosal organs as part of this mucosal defence. Meanwhile, Kozlov et al. (2011) shown that SACS mRNA expressed in nervous system included in brain and eye. The results are similar with our study, showed that maSACS transcript level was significantly detected in nervous-related system.
Fish larvae are exposed to pathogens a long period before of time until maturation of their immune systems, therefore information of immune gene may be essential for survival (Zapata et al., 1997). However, the mRNA expression analysis of immune related genes in loach is insufficient. Therefore, in this study, to information of RAG-1 and SACS mRNA expression, we analyzed the expression pattern of both genes during embryonic and early larval development. As a result, maRAG-1 mRNA was first detected in 0 dph, observed the increased mostly until 40 dph (Fig. 2). Significant expression of maRAG-1 was detected in 0 to 40 dph, while maSACS was detected in U to E3 and 0 to 40 dph (Fig. 3). In our study, the immune system of B and T cell marked by maRAG-1 starts to develop its function after hatching. The patterns of maSACS expression levels, they did not follow the trend of an initial increase after hatching. The detection of maSACS in unfertilized egg provides evidence of maternal contribution of these immune-relevant transcripts to the early embryo transcriptome. Rise et al. (2012) reported that expression profiled immune-relevant genes in Atlantic cod, the sacsin (SACS) have been expressed in unfertilized eggs and cleavage-stages embryos. Generally, maternal immunity plays very important role before immunelogical maturation. However, if maternal mRNA (or protein) ran out, this interval would be one crisis period. This finding may explain the reason of mass mortality breakout during the larval stage from immunological aspect.
In summary, this study provides the information of maRAG-1 and maSACS mRNA expression during the early developmental stage in loach. These expression patterns of both genes in tissue distribution and developmental stage shows that may play an important function in the early immune response of loach larvae. This study is the first to report the mRNA expression of the important immune-related genes, maRAG-1 and maSACS. Although it is still not clear whether presence of components of the immune system confer any protection against pathogens, maternal supply and early expression may indicate an important function during the early stages of development in loach.